Advances in Bioscience and Biotechnology
Vol.2 No.6(2011), Article ID:8903,6 pages DOI:10.4236/abb.2011.26066
Molecular investigation of the Arabian horse breeds of racing, productivity and longevity
![]()
1Biotechnology Department, Faculty of Science,Taif University, Taif, Saudi Arabia;
2Department of Biochemistry, Faculty of Veterinary Medicine, Minufiya University, Sadat City, Egypt;
3Department of Zoology, Faculty of Science, Cairo University, Cairo, Egypt;
4Economic Entomology and Pesticides Department, Faculty of Agriculture, Cairo University, Cairo, Egypt.
Email: *yasser92us@yahoo.com
Received 14 September 2011; revised 21 October 2011; accepted 9 November 2011.
Keywords: Arabian Horse; Thoroughbreds; Mitochondrial DNA; Longevity; Race; Productivity; Genetic Diversity
ABSTRACT
The Arabian; horse Equus caballus is belonging to family Equidae that all of its species are in danger of extinction. We sequenced 1059 nucleotides from three mitochondrial DNA protein coding genes for some breeds of the Arabian and English horses designated for race, productivity and longevity in order to investigate their genetic variation. The sequenced genes were NADH dehydrogenase subunit 2 (ND2), cytochrome C oxidase subunit 1 (CO1) and ATPase subunit 6 (ATP6). ATP6 discriminated among the different breeds where seven substitutions were recorded of which 2 were non-synonymous. The other loci recoded 8 synonymous substitutions of transitions most of which were among the breeds of longevity. The data of CO1 gene were used to construct the relationship of the Arabian horse to the thoroughbreds. Maximum-likelihood (ML) and Neighbor-joining (NJ) analytical methods were used. The two analytical methods showed clustering of the Arabian breeds in one group and the non-sister relationship within the thoroughbreds. One of the thoroughbreds clustered within the Arabian breeds suggesting the possibility of their out breeding. Increasing tendency in the variation of the studied genes was proportional to the utility of these genes in energy production within the mitochondria. The present study is therefore a preliminary step in the conservational efforts of the Arabian horse of equestrian.
1. INTRODUCTION
Arabian horse (Equus caballus) was believed to be one of the oldest and the most influential horse breeds in the world. Five major breeds of the Arabian horse are known as Keheilan, Seglawi, Abeyan, Hamdani and Hadban with heterogeneous origin [1]. Private farms in Saudi Arabia contain some of the above-mentioned breeds from which some breeds have been hybridized with European racing and performance breeds. Generally, the molecular studies on horse have three aims: the first is to know the genetic relationships between different breeds [1-9], the second is to know their origin [10-16] and the third is to investigate the genes of race, longevity and performance [17- 20].
It was believed that the characteristics of race efficiency of the horses are being inherited through the maternal but not the paternal genetics [18]. Genes of mitochondrial DNA play a role in biochemical processes associated with force, speed and performance efficiency [17] and have a role in cellular oxidation reactions required for vital activities. Harrison and Turrion-Gomez [18] studied eight genes of mitochondrial DNA, which have an effective role in the sportive efficiency of horses and proved that this genome is very important and is inherited to generations through the dam. The authors have revealed that different combinations of the genes of sportive efficiency may play a role in the efficiency of racehorses in terms of the race distance and age. A recent study revealed that there are 18 mitochondrial genes influence fitness and performance phenotypes [20]. The author revealed that these genetic data related to exercise suggest that there will be a great interest in studying equine genetics for performance evaluation and sports medicine applications.
The present study aimed to sequence and analyze mitochondrial DNA genes of some available horse breeds from Saudi Arabian private farms in order to investigate their relationship to other breeds such as thoroughbreds. As some molecular studies have supported the assumption that the characteristics of race efficiency are being inherited through the mitochondrial genome, the present study aimed also to discuss the genetic diversity between the breeds of race, productivity and longevity.
2. MATERIALS AND METHODS
We obtained 15 samples from Al-Massara International Equestrian Center. This center is reputed to be one of the largest horse riding centers in the Middle East and the Gulf region. It is located within the esteemed Alwasiliyyah district of Taif City. Blood samples were collected from four different breeds, three samples from each one as follow: Arabian horse for Longevity (Saqre, Nijm and Mubarak), English horse for race (Freyal, Ezz, Ahmar), Arabian horse for productivity (Qumrah, Nor Al-Jazeera, Jana) and English horse for productivity (Joy, Zahrah and Tabuk). With respect to the breed for race, samples were collected during rest state and after 15 minutes of exercise.
All blood samples were collected from jugular vein into heparinized tubes, labeled and immediately taken into the lab and stored at –80˚C until processing. DNA was extracted from 0.5ml blood samples with QIAGEN spin-column kits (QIAGEN Inc. Valencia, CA, USA) according to the manufacturer’s instruction. Extracted DNA was spectrophotmetrically quantified at 260 nm and DNA quality was determined by 260/280 nm ratio. DNA was used for polymerase chain reaction (PCR).
PCR was carried out in a 50 mL reaction mixture containing; 25 mL QIAGEN Taq PCR Master Mix Kit (Taq DNA Polymerase, PCR Buffer, MgCl2, ultrapure dNTPs at optimized concentrations), 2 mL DNA template, 0.2 mM as a final concentration of each primer and 21.6 mL autoclaved, distilled water to reach a total volume of 50 mL. The PCR cycling conditions for ND2 and AT6 genes were 30 cycles of denaturation at 94˚C for 30 s, annealing at 57˚C for 1 min and extension at 72˚C for 1 min. Similar PCR conditions were used for amplifying CO1 gene but with 40 cycles and annealing at 55˚C for 1 min. PCR products were analyzed by electrophoresis in a 1.5% agarose gel with ethidium bromide staining. To confirm the amplification of the targeted genes, the PCR products were excised after electrophoresis, purified using BioFlux BioSpin Gel Extraction kit (Bioer technology Co., Ltd) and sequenced. Sequencing reactions were performed in a MJ Research PTC-225 Peltier Thermal Cycler using an ABI PRISM. BigDyeTM Terminator Cycle Sequencing Kits with AmpliTaq-DNA polymerase (FS enzyme) (Applied Biosystems) according to the manufacturer’s protocol. Single-pass sequencing was performed on each template using the above-mentioned PCR-primers. The primers for PCR amplification and genes sequencing were designed on the basis of published sequences in the GenBank (NC_001640)The positions and sequences of the primers that have been used in both amplifications and sequencing are listed (Table 1).
Nucleotide sequences of the mitochondrial CO1 region (412 bp) were aligned with those obtained from the DDBJ database (4 E. caballus thoroughbred). One additional outgroup taxan (E. asinus) was included in the alignment in order to root the tree. The alignment was carried out by using the DNASIS 3.5 (Hitachi) and MacClade 4.03 (Sinauer Associates, Inc.) with manual adjustments. We conducted the tree analyses by neighborjoining (NJ) and maximum-likelihood (ML) methods. These analyses were done in PAUP* 4.0b10 [21] by heuristic searches with the TBR branch swapping, 10 random taxon additions and 1000 bootstrap replications for each method.
3. RESULTS AND DISCUSSION
Unambiguous 1059 sites from ND2, CO1 and ATP6 genes for 15 samples of the Arabian breeds of E. caballus for race, longevity and productivity were sequenced in this study. These data were deposited in DDBJ/EMBL Gen-Bank database with their accession numbers (AB663508- AB663552). The sequences were concatenated and the gab-containing sites were deleted so that 916 bp were left for analysis. The data showed base frequencies of A = 28.3%, C = 29.4%, G = 14.8% and T = 27.5%. Of these nucleotides, 899 were constant and 17 were variables. Six of the variable sites were parsimony-uninformative and 11 were informative under parsimony criterion. The estimated pairwise genetic distance (data not shown) was lower among the different haplotypes (D range = 0.0098 - 0.001) compared to that a these breeds and its congeneric E. asinus (D = 0.013). The estimated genetic distance showed a tendency to be high between the breeds for race and/or longevity and those for productivity and it was lower between those for race and longevity. Figure 1 depicts an NJ tree based on the nucleotide sequences of 412 unambiguous sites of CO1 gene aligned in 4 specimens of thoroughbred and 7 Arabian Equestrian breeds. An optimal ML tree was also found with similar topology and a negative log likelihood of 816.2.
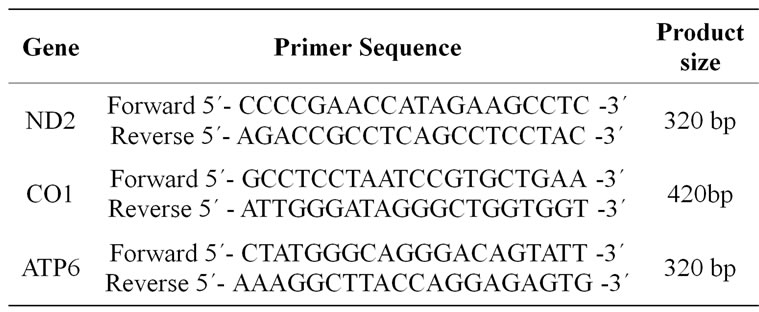
Table 1. Primers used for PCR amplification and sequencing of horse genes.
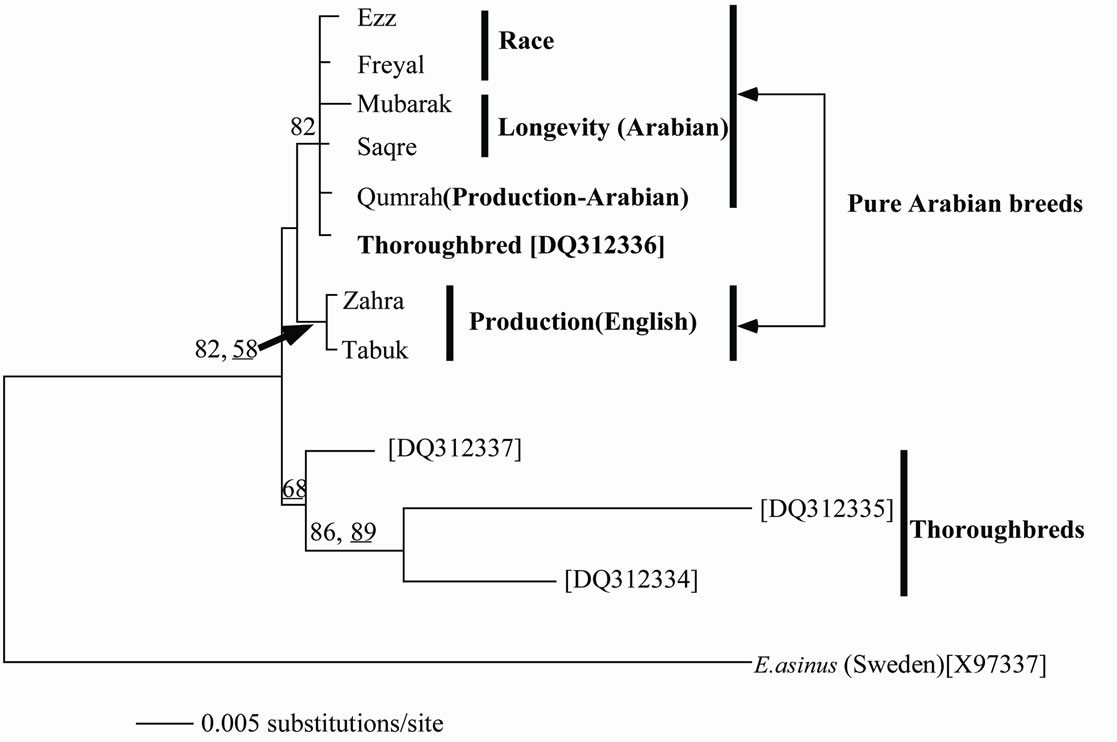
Figure 1. Neighbor-joining tree constructed from 412 bp of CO1 gene among the different studied breeds. The underlined values at nodes designate the bootstrap values when the maximum-likelihood method was conducted, while the other values represent the NJ bootstrap support.
The overall base compositions of the L-strand as accounted by the ML analysis were A: 26.5.8%; T: 29.2.4%; C: 26.4% and G: 17.9%. The two analytical methods (NJ and ML) showed clustering of the Arabian breeds (those for race, longevity and productivity in one group. This clustering was trustable because of the reasonably strong statistical support (BP = 82). One of the thoroughbreds clustered within the Arabian breeds while the other thoroughbreds showed sister group relationship to the Arabian group suggesting the impurity of these Arabian breeds. The English breeds for productivity were found in a separate cluster within the Arabian group with reasonable statistical support (BP = 82 and 58). A study on the polymorphism of RAPD-PCR and D-loop of the mtDNA supported distant relationships of the Arabian breeds to the thoroughbreds [8].
Nucleotide substitutions are generally considered in terms of changes within the two structural classes of nucleotides (purines and pyrimidines), that is, in terms of transitions and transversions. 320 bp from ND2 gene showed only one synonymous base transition (T to C) in the race samples and in one of the longevity samples (data not shown). For the barcoding gene codes for cytochrome C oxidase, the sequenced 413 bp showed 7 synonymous transitions (A to G and C to T) among the samples from the longevity breeds only (Figure 2). This fragment showed 29 substitutions (11 transversions and 18 transitions) between the studied haplotypes and the thoroughbreds. Among these mutations, 18 were in the third position and 11 were in the first and second positions and therefore, thirteen substitutions were non-synonymous and 16 were synonymous. One of these thoroughbreds showed complete amino acid identities to the present breeds (data not shown).
ATP6 gene showed the greatest variations among the different samples studied (Figure 3) and therefore, it could be considered as an important marker to study the possible polymorphisms in the Arabian E. caballus breeds. The polymorphism of the 320 base pair representing the partial sequence of the ATP6 gene in 15 individuals of Arabian E. caballus was analyzed. Seven polymorphic sites in this fragment defined four haplotypes. These sites included six transitions and one transversion, which accounted for five synonymous and two nonsynonymous substitutions. The non-synonymous changes were found in the breeds of productivity and involved a substitution of glycine with arginine at G331➝A331 and a substitution of glutamine with arginine at A392➝G392 (Figure 3). This substitution may be interpretive with that arginine reduces lactic acid production by the inhibition of glycolysis and enhances work capacity and exercise tolerance [22].
The English Thoroughbred is the best known breed of horse in the western world. Thoroughbreds were developed during the seventeenth and eighteenth centuries in England, largely owing to the enthusiasm of English aristocracy for horse racing and betting [12]. Until recently, it was well-known that the paternal origins of these breeds are well documented as being derived from a handful of Middle Eastern stallions, the most influential

Figure 2. The aligned nucleotides of the sequenced fragment of CO1 gene in the different breeds as follow: Tabuk (A), Joy (B), Jana (C), Nor Al-Jazeera (D), Qumrah (E), Saqre (F), Nijm (G), Mubarak (H), Freyal (I) and Ahmar (J). The underlined columns are those with polymorphism.
of which are Godolphin Arabian, Darley Arabian and Byerley Turk [15]. A further study proved a similar hypothesis that the Arab breed is one of the most influential horse breeds in the World [10]. It is distributed worldwide and has been involved in the formation of many other horse breeds, such as the Thoroughbred [11] or the Lipizzan [13]. An up to date molecular study argued strongly against this claim. Bower et al. [16] stated that there is no evidence to support exclusive Arab maternal origins or a significant importation of Oriental mares (the term used in historic records to refer to Middle East and western Asian breeds including Arab, Akhal-Teke, Barb and Caspian). Instead, they showed that Thoroughbred foundation mares had a cosmopolitan European heritage with a far greater contribution from British and Irish Native mares than previously recognized. As supportive evidence to this hypothesis, Harrison and Turrion-Gomez [18] have stated that The thoroughbred is a composite breed and the various maternal lines have diverse geographical and founder breed origins established prior to the formation of the thoroughbred studbook. Using RAPDPCR markers, Mahrous et al. [8] revealed a high genetic distance between the Arabian breeds and the Thoroughbred supporting this postulation. The non-sister relationship within the Thoroughbreds in our tree indicated that some of the tested breeds are, perhaps, not purely Arabian, even they are of the best known breeds. This evidence supports the conclusion that many of the Arabian breeds are now mixed with the European thoroughbreds in order to improve their racing performance.
It has been postulated by Harrison and Turrion-Gomez [18] that the mitochondrial gene variation may represent a measurable component contributing to performance variability. The authors have proven this postulation in terms of the existence of D-loop variation in the populations of thoroughbreds. In the present study, increasing tendency in the variation of the studied genes was proportional to the utility of these genes in energy produc-

Figure 3. The aligned nucleotides of the sequenced fragment of ATP6 gene in the different breeds as follow: Tabuk (A), Joy (B), Jana (C), Nor Al-Jazeera (D), Qumrah (E), Saqre (F), Nijm (G), Mubarak (H), Freyal (I) and Ahmar (J). The underlined columns are those with polymorphism.
tion through the oxidative process within the mitochondria. ATP6, therefore showed the highest variability followed by CO1 gene. The least variation among the breeds was shown in ND2 gene.
4. CONCLUSIONS
In conclusion, the present study showed that the genetic structure of the different Arabian breeds is a preliminary guide for conservation management of Equus caballus. A wide-range survey to elucidate inter-and intra-breeds genetic diversity is necessary because: these breeds play a role in the global economy and the greater part of them is distributing in the developing countries. They are free from the genes of other foreign breeds, however, many breeds are found in Saudi Arabia as a result of cross breeding with European breeds specially, in the private farms.
5. ACKNOWLEDGEMENTS
This work has been done under the financial support of Taif University of Kingdom of Saudi Arabia (Project number 1/432/1350).
REFERENCES
- Głażewska, I. (2010) Speculations on the origin of the Arabian horse breed. Livestock Science, 129, 49-55. doi:10.1016/j.livsci.2009.12.009
- Caetano, A.R., Shiue, Y-L., Lyons, L.A., Brien, S.J.O., Laughlin, T.F., Bowling, A.T. and Murray, J.D. (1999) A comparative gene map of the horse (Equus caballus). Genome Research, 12, 1239-1249. doi:10.1101/gr.9.12.1239
- Mirol, P.M., García, P.P. and Dulout, F.N. (2002) Mitochondrial variability in the D-loop of four equine breeds shown by PCR-SSCP analysis. Genetics and Molecular Biology, 25, 25-28. doi:10.1590/S1415-47572002000100006
- Georgescu, S.E., Condac, E., Rebedea, M., DumitruTesio, C., Dinischiotu, A. and Costache, M. (2005) Arabian horses genotyping using seventeen microsatellites. Archiva Zootechnica, 8, 169-175.
- Glowatzki-Mullis, M.L., Muntwyler, J., Pfister, W., Marti, E., Rieder, S., Poncet, P. A. and Gaillard, C. (2005) Genetic diversity among horse populations with a special focus on the Franches-Montagnes breed. Animal Genetics, 37, 33-39. doi:10.1111/j.1365-2052.2005.01376.x
- Głażewska, I., Wysocka, A., Gralak, B., Prus, R. and Sell, J. (2007) A new view on dam lines in Polish Arabian horses based on mtDNA analysis. Genetics Selection Evolution, 39, 609-619. doi:10.1186/1297-9686-39-5-609
- Cervantes, I., Molina, A., Goyache, F., Gutiérrez, J.P. and Valera, M. (2008) Population history and genetic variability in the Spanish Arab Horse assessed via pedigree analysis. Livestock Science, 113, 24-33. doi:10.1016/j.livsci.2007.02.011
- Mahrous, K.F., Alam, S.S. and Hassan, A.M. (2010) Genetic variations between horse breeds using RAPD markers. Nature and Science, 8, 90-99.
- Cieslak, M., Pruvost, M., Benecke, N., Hofreiter, M., Morales A., Reissmann, M. and Ludwig, A. (2010) Origin and history of mitochondrial DNA lineages in domestic horses. PLoS One, 5, e15311. doi:10.1371/journal.pone.0015311
- Bowling, A.T. and Ruvinsky, A. (2000) The genetic resources and their conservation. The Genetic of the Horse. CAB International, Wallingford.
- Bowling, A.T. and Ruvinsky, A. (2000) Genetic aspects of domestication. The Genetic of the Horse. CAB International, Wallingford.
- Cassidy, R. (2002) The sport of kings: Kinship, class and thoroughbred breeding in Newmarket. Cambridge University Press, Cambridge. doi:10.1017/CBO9780511613760
- Zechner, P., Sölkner, J., Bodo, I., Druml, T., Baumung, R., Achmann, R., Marti, E., Habe, F. and Brem, G. (2002) Analysis diversity and population structure in the Lipizzan horse bred based on pedigree information. Livestock Production Science, 77, 137-146. doi:10.1016/S0301-6226(02)00079-9
- Royo L.J., Álvarez, I., Beja-Pereira, A., Molina, A., Fernández, I., Jordana, J., Go´mez, E., Gómez, J.P. and Goyache, F. (2005) The origins of the Iberian horses assessed via mitochondrial DNA. Journal of Heredity, 96, 663-669. doi:10.1093/jhered/esi116
- Hewitt, A. (2006) Sire lines. Eclipse Press, Lexington.
- Bower, M.A., Campana, M.G., Whitten, M., Edwards, C.J., Jones, H., Barrett, E., Cassidy, R., Nisbet, R.E.R., Hill, E.W., Howe, C.J. and Binns, M. (2011) The cosmopolitan maternal heritage of the Thoroughbred racehorse breed shows a significant contribution from British and Irish native mares. Biological Letters, 7, 316-320. doi:10.1098/rsbl.2010.0800
- Lesage, R., Simoneau, J.A., Jobin, J., Leblanc, J. and Bouchard, C. (1985) Familial resemblance in maximal heart rate, blood lactate and aerobic power. Human Heredity, 35, 182-189. doi:10.1159/000153540
- Harrison, S.P. and Turrion-Gomez, J.L. (2006) Mitochondrial DNA: An important female contribution to thoroughbred racehorse performance. Mitochondrion, 6, 53-66. doi:10.1016/j.mito.2006.01.002
- Sobczyńska, M. (2006) Genetic correlations between racing performance at different racing distances in Thoroughbreds and Arab horses. Czech Journal of Animal Science, 51, 523-528.
- Barrey, E. (2010) Genetics and genomics in equine exercise physiology: An overview of the new applications of molecular biology as positive and negative markers of performance and Health. Equine veterinary Journal, 42, 561-568. doi:10.1111/j.2042-3306.2010.00299.x
- Swofford, D.L. (2002) PAUP*: Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10, Sunderland, Sinauer Associates, Inc., Publishers, Sunderland,
- Burtscher, M., Brunner, F., Faulhaber, M., Barbara Hotter, B. and Likar, R. (2005) The prolonged intake of Larginine-L-aspartate reduces blood lactate accumulation and oxygen consumption during submaximal exercise. Journal of Sports Science and Medicine, 4, 314-322.

