Advances in Bioscience and Biotechnology
Vol.1 No.4(2010), Article ID:2861,8 pages DOI:10.4236/abb.2010.14043
Sponge-associated bacteria of Lakshadweep coral reefs, India: resource for extracellular hydrolytic enzymes
![]()
1Microbiology Laboratory, National Institute of Oceanography-Regional Centre, Kochi, India;
2Microbiology Laboratory, National Institute of Oceanography, Goa, India.
Email: shanta@nio.org
Received 8 July 2010; revised 19 July 2010; accepted 31 July 2010.
Keywords: Sponge-Associated Bacteria; Extracellular Hydrolytic Enzyme; Coral Reef; Lakshadweep
ABSTRACT
Sponges (Phylum: Porifera) is one of the major groups in the Lakshadweep coral reefs. These sponges harbor diverse bacteria with metabolic potentiality. From biodiversity to biotechnological prospecting, scientific investigations related to sponge associated microorganisms have expanded, but remain rather limited to few geographic locations. In this study, culturable bacteria associated with two demosponges viz Dysidea granulosa, Sigmadocia fibulata and the ambient water were screened for commercially important enzymes such as amylase, protease, gelatinase, lipase, deoxyribonucleic, phosphatase and urease. Amylase and phosphatase were the predominant enzymes produced by >80% of sponge-associated bacteria compared to the ambient water. Nearly 50% of the sponge-associated bacteria expressed multiple enzymatic activities (> 4) with variation in the percentage of expression of individual enzymes. More than 65% of the culturable heterotrophic bacteria associated with sponges were Gammaproteobacteria. The order Vibrionales was the main source for multiple enzyme production. Sponge associated bacteria formed more closely related clusters than the water isolates based upon their activity pattern. High recovery of sponge-associated bacteria with multiple enzymatic activities suggest that these versatile bacteria are yet unexploited potential for bioprospecting.
1. INTRODUCTION
Coral reefs are highly productive natural ecosystem and provide an excellent habitat for a vast array of marine organisms due to their structure, efficient biological recycling and high retention of nutrients [1]. Class Demospongiae comprises about 85% of the living sponges and is a major component of benthic community in the coral reef ecosystem [2]. Sponges filter 24,000 L of sea water kg-1 day-1, resulting in colonization of large number of extracellular bacteria on the surface and internal mesohyl matrix. In addition, these unique ecological niches have copious amount of particulate organic matter [3,4]. A single sponge host can be inhabited by diverse bacteria. Bacteria which constitute 40% of the sponge volume [5] produce extracellular hydrolytic enzymes that facilitate the metabolism of complex organic matter thereby assisting the host in nutrition and various metabolic processes. Bacterial enzymes provide a greater diversity of catalytic activity and can be produced economically. Currently bacteria from terrestrial sources are employed for industrial production of enzymes. Sponges are considered as microbial fermenters that provide exciting new avenues in marine microbiology and biotechnology [6]. Although, the potentialities of hydrolytic enzymes from marine bacteria have been recognized, studies on enzymes from bacteria associated within the microhabitats of sponge for biotechnological application are rare [7]. We hypothesize that sponge associated bacteria are excellent source of not only enzymes but also multiple extracellular hydrolytic enzymes than seawater bacteria due their characteristic nutrient rich microhabitat provided by the host organism. The present study is the first of its kind which focuses on bioprospecting on various degrading enzymes from culturable heterotrophic marine bacteria associated with two sponges viz. Dysidea granulosa and Sigmadocia fibulata and ambient seawater in the Lakshadweep Islands. These bacteria can be effectively cultivated in culture media for production of enzymes.
2. MATERIALS AND METHODS
2.1. Sample Collection
In this study, we examined the bacterial composition of two sponges, Dysidea granulosa Bergquist, 1965 and Sigmadocia (Haliclona) fibulata Schmidt, 1862 (here after mentioned in this article as Sigmadocia fibulata) belongs to Phylum: Porifera; class: Demospongiae; subclass: Ceractinomorpha. D. granulosa belongs to the order Dictyoceratida and S. fibulata belongs to the order Haplosclerida. Sponges and ambient seawater were collected from two locations, one outside the lagoon (10° 34’ 25.64” N; 72° 38’ 57.48” E) and the other inside the lagoon (10° 34’ 44.51” N; 72° 38’ 24.11” E) of Kavaratti Island, Lakshadweep, west coast of India (Figure 1) by SCUBA diving. These two species of sponges are very distinctive in gross morphology and microhabitat. Sponges were transferred immediately to Whirlpak sterile sampling bags (Nasco) and sealed underwater to prevent contact with air and possible oxidation and contamination. Ambient seawater was collected using 1.8L Niskin water sampler within 1m of the sponge colony and transferred to Whirlpak sterile sampling bags. Samples were processed within 2-3 hours of collection.
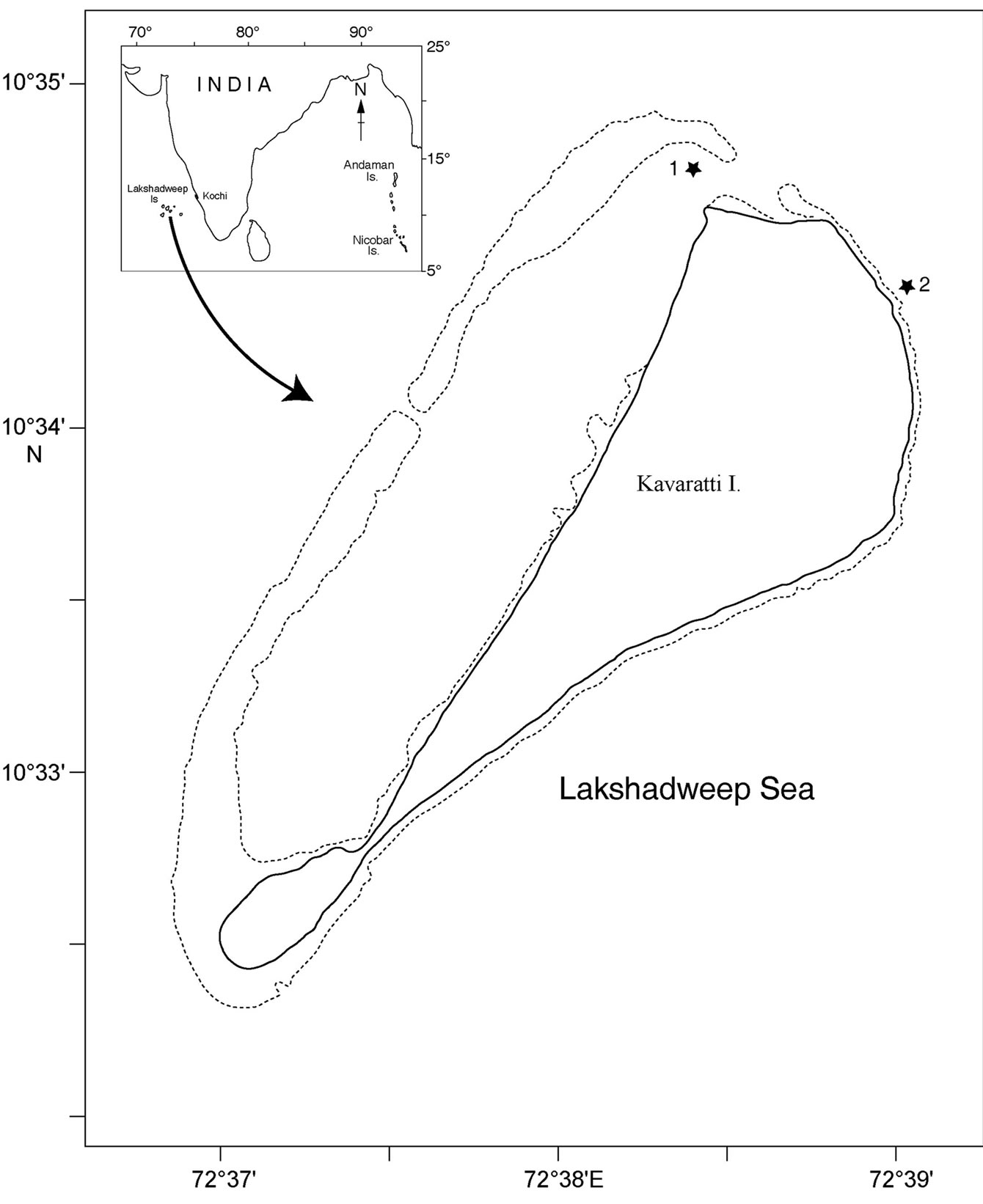
Figure 1. Map of Kavaratti Island, India showing sampling site 1 (inside the lagoon) and 2 (out side the lagoon).
2.2. Bacterial Isolation
Sponges were washed thoroughly with autoclaved; filter sterilized (0.22 µm-pore size filter) seawater until visibly free of debris and sediments. They were then homogenized using sterile mortar and pestle. Serially diluted sponge homogenate and direct sub samples of ambient water were spread plated onto various media differing in composition and concentration. Zobell marine agar was prepared in distilled water, nutrient broth of varying strength (100%, 50%, 25%, 10%) supplemented with 1.5% agar, OLIGO media (0.005% yeast extract, 0.05% tryptone, 0.01% sodium glycerophosphate, 1.2% Noble agar) prepared in seawater [8] were the different media employed for bacterial isolation. All plates were incubated at 28°C for 7 days. Random selection allowed a comprehensive collection of culturable heterotrophic bacteria found in this ecosystem. Pure cultures were cryopreserved in glycerol broth at –80°C. Bacterial isolates were identified using standard methods described [9,10].
2.3. Detection of Enzymatic Activity
Bacteria were screened for various hydrolytic enzymes such as amylase, protease, deoxyribonuclease, gelatinase, lipase, phosphatase and urease. Pure cultures were inoculated on plates supplemented with respective substrates and incubated at 28°C for 3 days and presence or absence of enzymatic activity were detected using the methods described below.
2.3.1. Amylase
Nutrient agar supplemented with 0.2% soluble starch were used to screen for amylase production. After incubation the plate was flooded with lugol’s iodine solution. Presence of clear zone around the culture was taken as positive result and formation of blue color in the medium surrounding the colony was the negative reaction.
2.3.2. Protease
Skim milk (5%, final concentration) incorporated in nutrient agar was used as the screening medium. Clear zone around the bacterial colony after incubation indicated hydrolysis of casein, which is the positive reaction. No zone around the colony indicates negative reaction.
2.3.3. Deoxyribonuclease
DNase test agar base (Himedia, India) contains 0.2% DNA in final concentration (wt/wt). After incubation, DNase plate was flooded with 0.1% toluidine blue solution. Blue characteristic precipitate formed on bacterial colony is the positive reaction.
2.3.4. Gelatinase
Incorporated 0.4% gelatin (wt/wt) in nutrient agar and flooded with 15% mercuric chloride (wt/v) in concentrated hydrochloric acid (v/v) after incubation. Gelatinase positive isolates exhibits a clear zone around the colony.
2.3.5. Lipase
Nutrient agar was supplemented with 0.01% (w/v) of calcium chloride and 1% of Tween 80 (v/v) (polyoxyethylene sorbitan monooleate). After incubation presence of halo around the colony is regarded as positive reaction by the formation of calcium soaps with oleic acid. Absence of halo demarcates negative reaction.
2.3.6. Phosphatase
Phenolphthalein phosphate agar containing phenolphthalein diphosphate as phosphate source were inoculated and exposed to ammonia vapor after incubation period. Positive isolates turns into pink colored colonies after exposure.
2.3.7. Urease
Christensen agar base were supplemented with 40% (w/v) urea solution were inoculated and incubated. A change in the colour of the yellowish medium to a pink coloration around the bacterial colony is regarded as positive reaction.
2.3.8. API ZYM Micro Method
Selected candidate isolates with promising enzymatic activity were semi quantitatively determined with API ZYM (API bioMerieux Ltd) micro method [11]. The procedure was as per the manufactures instruction and expressed in nm of hydrolyzed substrates.
2.4. Statistical Analysis
Presence or absence data were square root transformed prior to analysis. Bray-Curtis similarities were used to produce a similarity matrix based on presence of enzyme (indicated by 1) or absence (indicated by 0). For the construction of a dendrogram demarcating the similarity of activity pattern of the bacterial isolates, group average linkage in the hierarchical clustering algorithm was performed using the PRIMER v6.1 program [12].
3. RESULTS
3.1. Identification of Heterotrophic Culturable Bacteria
Culturable bacteria associated with S. fibulata and D. granulosa mostly belonged to Gammaproteobacteria followed by Firmicutes, Actinobacteria and Betaproteobacteria. Ambient water isolates were dominated by Actinobacteria followed by Gammaproteobacteria, Firmicutes and Betaproteobacteria. Betaproteobacteria were absent in D. granulosa (Table 1). Although the two sponges vary considerably in ecological distribution and

Table 1. Percentage of culturable heterotrophic bacterial groups of D. granulosa and S. fibulata and ambient seawater.
conditions, bacterial isolates reflected a striking similarity at the phylum level in their microbial communities. Majority of the sponge-associated Gammaproteobacteria belonged to the order Vibrionales.
3.2. Extra Cellular Enzymatic Activity
Bacterial isolates showed activity for at least one enzyme screened irrespective of their location and source. Amylase and phosphatase were the dominant enzymes in this system. Urease was the least expressed enzyme. Sponge associated bacteria exhibited higher percentage of hydrolytic enzymatic activity compared to the bacteria from ambient water (Figure 2). Though all the enzymes were expressed by both species, there was considerable variation in the percentage of positive isolates. Occurrence of hydrolytic enzymes such as amylase, lipase and protease was higher in. D. granulosa isolates than in S. fibulata (Figure 3). However, phosphatase and Deoxyribonuclease activity were higher in the case of S. fibulata (89.1% & 47.8%) than in D. granulosa (85% & 29.9%). Manifestation of multiple enzymatic activities was observed in bacteria associated with sponge (Figure 4). Nearly 50% of the sponge associated culturable bacteria expressed > 4 hydrolytic enzymes, where as only 33% of the ambient water isolates could produce at least 2 enzymes. Selected candidate isolates showed promising enzymatic activity using API ZYM. The activity of the extra cellular hydrolytic enzymes of the bacteria is shown in Figure 5. All the isolates showed higher phosphatase activity. Ambient water isolates showed relatively lower lipase activity. The bacteria associated with the sponge D. granulosa showed the highest activity for all enzymes tested.
3.3. Statistical Analysis
Based upon the enzymatic activity pattern bacterial isolates were grouped into clusters using Bray–Curtis similarity matrix. Bacterial isolates from both sponges show a high similarity. They form 26 clusters (Figure 6) and the lowest similarity among the bacterial isolates from sponges were 40.3% whereas the water isolates formed
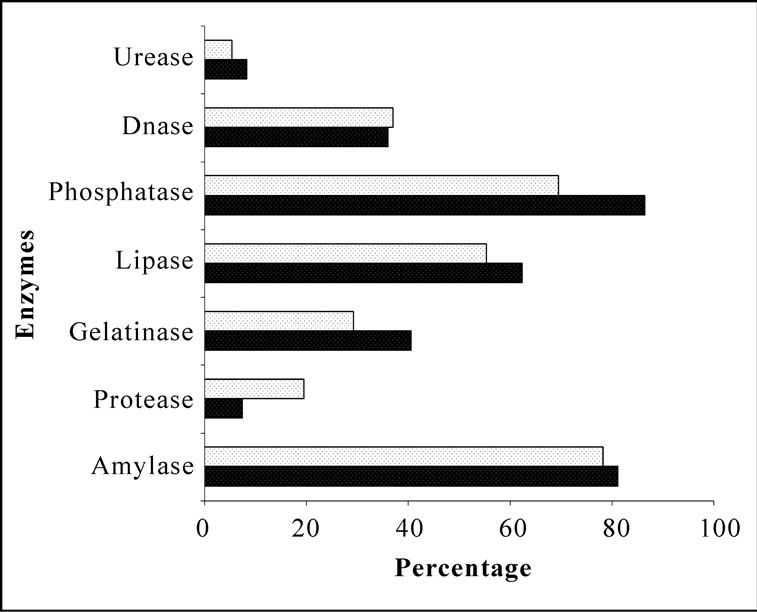
Figure 2. Comparison of percentages of heterotrophic bacterial isolates of ambient seawater and sponges with extracellular hydrolytic enzymes.  Water;
Water;  Sponge.
Sponge.
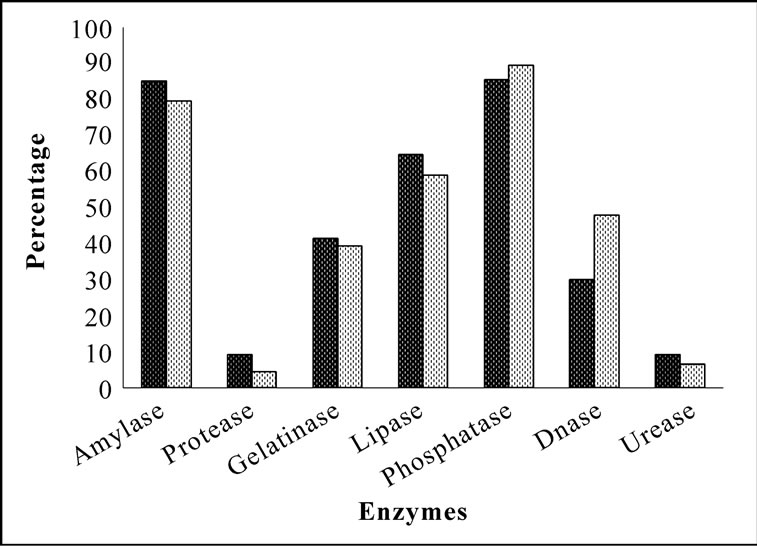
Figure 3. Comparison of percentage of culturable heterotrophic bacteria with extracellular hydrolytic enzymatic activity in D. granulosa and S. fibulata.  D. granulosa;
D. granulosa;  S. fibulata.
S. fibulata.

Figure 4. Frequency of incidence of multiple extracellular hydrolytic enzymes in bacteria associated with sponges and ambient seawater.  D. granulosa;
D. granulosa;  S. fibulata;
S. fibulata;  Water.
Water.

Figure 5. Semi quantification of potential isolates with multiple enzymatic activities from different sources using API ZYM.  Phosphatase;
Phosphatase;  Lipase;
Lipase;  Protease;
Protease;  Gelatinase.
Gelatinase.

Figure 6. Cluster analysis showing the similarity (%) of activity pattern among sponge isolates. Dysidea granulosa indicated as S-DG and Sigmadocia fibulata as S-SF. Total isolates showing the same activity pattern is given in parentheses.
34 clusters and the lowest similarity of 16.5% was recorded among water isolates (Figure 7).
4. DISCUSSION
Microorganisms play a central role in sponge biology, as they are associated with many sponges either extracellularly, intracellularly or both. They also serve as food source for the host. Isolation is a mandatory approach to obtain novel microbes and also for evaluating their biochemical characteristics to understand the ecophysiological and environmental functions vis a vis their potential applications [13]. The discovery of enormous microbial diversity in marine sponges provides unprecedented research opportunities. Increased metabolic capabilities of sponge-associated bacteria were directly correlated with increased levels of potentially available nutrients in the sponge. Sponges filter seawater and accumulate copious amount of organic matter within the choanocytic chambers along with bacteria [8,14]. Transfer of photosynthate by the cyanobacterial symbionts to the sponge was proven [15], likewise the abundant bacterial population within the mesohyl, choanocytes and aquiferous chambers were also actively participating in the sponge nutrition by secreting extracellular enzymes and it acts upon the particulate organic matter accumulated within the sponge body. Although some dissolved inorganic and organic components of small size can be more or less directly absorbed by the organisms, many of the vital carbonand/or nitrogen-containing compounds are in the form of macromolecular structures in the natural environment. To access these substrates, microorganisms must secrete enzymes capable of hydrolyzing these compounds [16]. In the presence of vast amount of substrates, bacteria are induced to produce extra cellular enzymes and release them into immediate surroundings leading to the degradation of various complex molecules accumulated. Consequently, the released nutrients can be taken up by the sponge [17,18]. Seawater isolates were limited in their ability to utilize different nutritional sources and appeared to be metabolically restricted due to the reduced concentration of nutrients in the water column [19]. Amylase was the most prominent enzyme in the study region. High amylase activity has also been reported from bacteria associated
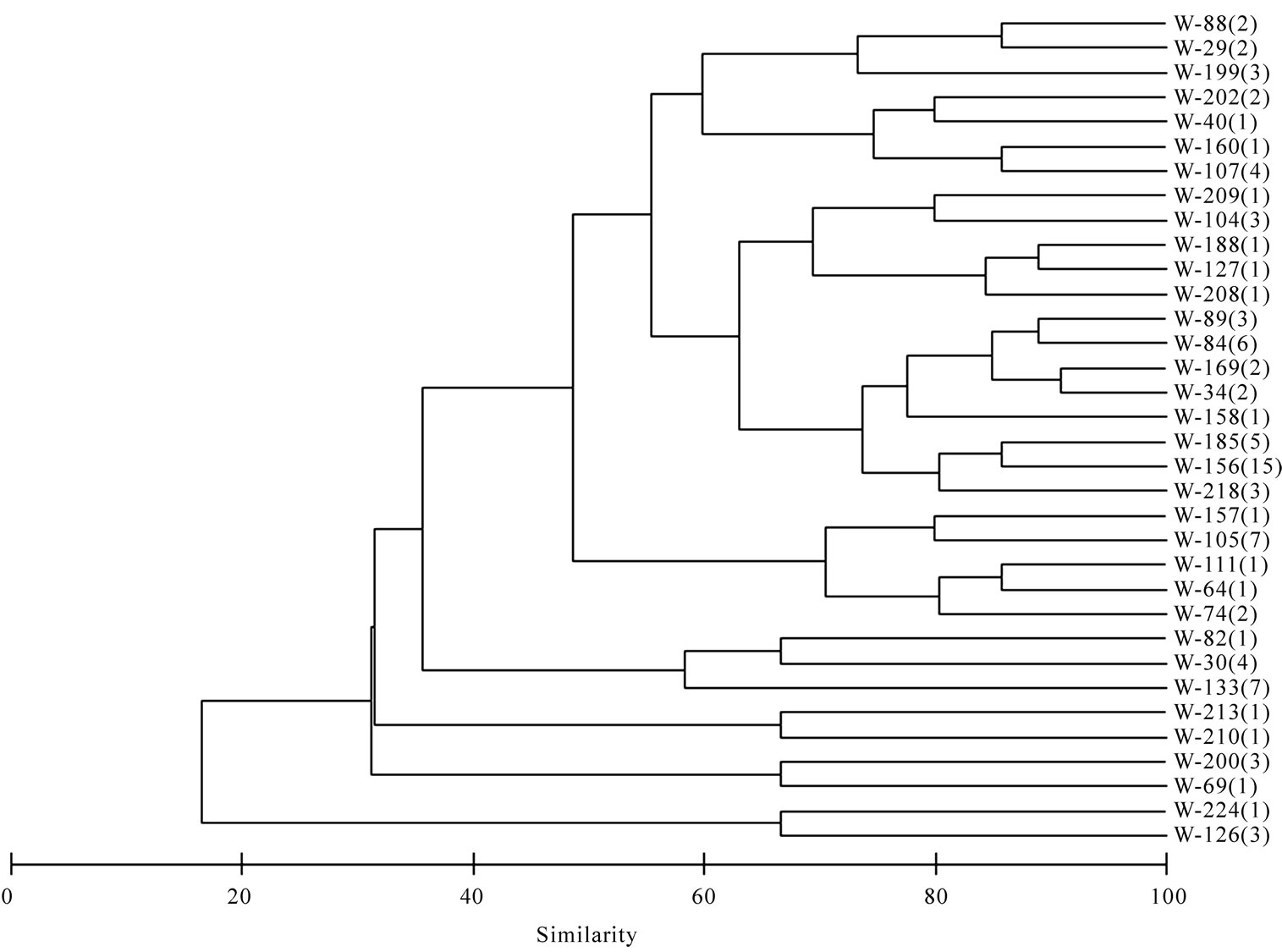
Figure 7. Cluster analysis showing the similarity (%) of activity pattern among water isolates. Total isolates showing the same activity pattern is given in parentheses.
with marine sponge Fasciospongia cavernosa, collected from the eastern peninsular coast of India [20]. Hydrolytic enzymes such as lipases still belong to the most important biocatalysts, because they accept a broad range of substrates, and are usually very stable in organic solvents [21]. The ability of sponge-associated bacteria to produce other hydrolytic enzymes such as phosphatase, lipase and gelatinase suggests that they play an important role in the degradation of organic matter and help in regeneration of nutrients. Most of the bacteria associated with the Caribbean demosponge Ceratoporella nicholsoni were able to utilize substrates like DNA, gelatin and a range of fatty acids like Tween 40 to Tween 80 [19]. Between the two species presently studied, differences were observed in the number of isolates and their enzyme activity. This could be due to the differences in their physiological requirement and the ambient variation in the quality and quantity of the organic matter. Associated bacteria of the marine sponge Ircinia dendroides from the Mediterranean Sea, multiple enzyme expression of gelatinase and lipase were observed [22].
In the present study the highest percentage (89%) of sponge-associated bacteria showed phosphatase activity. Hauksson [23] have described phosphatase producing marine vibrio sp. Bacteria associated with the marine sponge Haliclona sp have also been reported producing phosphatase enzyme [24]. Zaccone [25] opinioned that bacteria play a key role in the P cycle and that phosphatase was widespread in marine bacteria. Some bacteria associated with D. granulosa were found to produce urease enzyme in our study. Based on this study and earlier studies [19,20,24,26,27], it can be deduced that sponge-associated bacteria represent excellent sources for extracellular enzymes like amylase, protease, gelatinase, lipase, deoxyribonuclease and phosphatase. Enzymes from these bacteria can be obtained by mass fermentation followed by extraction and subsequent purification [20]. Further our results re-affirm that spongeassociated bacteria differ largely from the surrounding population of ambient seawater, both quantitatively and qualitatively.
Even though different media of varying compositions and concentrations have been used for isolation of heterotrophic bacteria, approximately 78% of the culturable bacteria associated with the sponge C. nicholsoni demonstrated phenotypic characteristics most closely related to species belonging to the orders Vibrionales and Aeromonadales [8]. From our study, Vibrionales emerged as the influential group inhabiting in both the sponges; they were abundant among the culturable bacteria as well as functionally dynamic with multiple enzymatic activities.
The present study has revealed the presence of high numbers of diverse culturable heterotrophic bacteria in association with sponges producing extracellular enzymes responsible for biopolymer degradation. D. granulosa is an excellent source of bacteria that can produce amylase, protease, lipase and urease, whereas S. fibulata associated bacteria produce phosphatase and deoxyribonuclease. The data obtained through the study reveals the crucial role played by culturable heterotrophic bacteria associated with sponge and seawater in the degradation of biopolymers. These kind of broad screening investigations indicate that the production of certain enzymes is not restricted to isolates belong to single source. It also improves our understanding of the functional role of bacteria in coral reef ecosystem. However, future studies with radio labeled and/or genetic probes are warranted to elucidate the action of bacterial enzymes in sponge nutrition. In particular, the development of new and innovative cultivation strategies holds great potential for accessing the microbial lineages that are so far under represented in culture. Mass fermentation of sponge–associated bacteria can provide a renewable resource of enzymes and harnessing these microbes for other metabolites as well as conserving the natural population of sponges.
5. ACKNOWLEDGEMENTS
The authors thank the Director, National Institute of Oceanography. AF thankfully acknowledges Council for Scientific and Industrial Research (CSIR) for the award of junior research fellowship. The laboratory facility was extended by MMRF of NIO, RC –Kochi funded by the Ministry of Earth Sciences, New Delhi. We thank taxonomist Dr. P. A. Thomas, Scientist Emeritus, CMFRI for identification of sponges. We also acknowledge Dr. C. T. Achuthankutty (visiting scientist, NCAOR, Goa) for critically reading the manuscript. This is NIO contribution number 4807.
REFERENCES
- Kleypas, J.A. (1997) Modeled estimates of global reef habitat and carbonate production since the last glacial maximum. Paleoceanography, 12(4), 533-545.
- Hooper, J.N.A. and van Soest, R.W.M. (2002). Phylum Porifera. Class Demospongiae Sollas, 1885 Kluwer Academic/ Plenum Publishers, New York.
- Vogel, S. (1977) Current-induced flow through living sponges in nature. Proceedings of the National Academy of Sciences of the United States of America, 74(5), 2069- 2071.
- Wehrl, M., Steinert, M. and Hentschel, U. (2007) Bacterial uptake by the marine sponge. Aplysina aerophoba. Microbial Ecology, 53(2), 355-365.
- Wilkinson, C.R. (1978) Microbial associations in sponges. II. Numerical analysis of sponge and water bacterial populations. Marine Biology, 49(2), 169-176.
- Hentschel, U., Usher, K.M. and Taylor, M.W. (2006) Marine sponges as microbial fermenters. FEMS Microbiology Ecology, 55(2), 167-177.
- Wang, G.Y. (2006) Diversity and biotechnological potential of the sponge-associated microbial consortia. Journal of Industrial Microbiology and Biotechnology, 33(7), 545-551.
- Santavy, D.L., Willenz, P. and Colwell, R.R. (1990) Phenotypic study of bacteria associated with the Caribbean sclerosponge, Ceratoporella nicholsoni. Applied and Environmental Microbiology, 56(6), 1750-1762.
- Holt, J.G., Krieg, N.R., Sneath, P.H.A., Staley, J.T. and Williams, S.T. (2000). Bergey's manual of determinative bacteriology. Lippincott Williams & Wilkins Philadelphia, USA.
- Gerhardt, P., Murray, R.G.E., Costilov, R.N., Nester, E.W., Wood, W.A., Kreig, N.R. and Philip, G.B. (1981) Manual of methods for general bacteriology. In: Philips, G.B. Ed., American Society for Microbiology, Washington DC, 409-443.
- Zdanowski, M.K. and Figuerias, F.G. (1999) CFU bacterial fraction in the esturine upwelling ecosystem of Ria de Vigo, Spain: variability in abundance and their ecophysiological description. Marine Ecology Progress Series, 182, 1-15.
- Clarke, K.R., Warwick, R.M., Somerfield, P.J. and Gorley, R.N. (2005) Methods manual: Changes in marine communities. PRIMER, 6(1), 10.
- Sfanos, K., Harmody, D., Dang, P., Ledger, A., Pomponi, S., McCarthy, P. and Lopez, J. (2005) A molecular systematic survey of cultured microbial associates of deep-water marine invertebrates. Systematic and Applied Microbiology, 28(3), 242-264.
- Wilkinson, C.R., Garrone, R. and Vacelet, J. (1984) Marine sponges discriminate between food bacteria and bacterial symbionts: Electron microscope radioautography and in situ evidence. Proceedings of the Royal Society of London, Series B-Biological Sciences, 220(1221), 519-528.
- Sara, M. (1971) Ultrastructural aspects of the symbiosis between two species of the genus Aphanocapsa (Cyanophyceae) and Ircinia variabilis (Demospongiae). Marine Biology, 11(3), 214- 221.
- Marx, J.C., Collins, T., D'Amico, S., Feller, G. and Gerday, C. (2007) Cold-adapted enzymes from marine antarctic microorganisms. Marine Biotechnology, 9(3), 293-304.
- Lee, Y.K., Lee, J.H. and Lee, H.K. (2001) Microbial symbiosis in marine sponges. Journal of Microbiology, 39(4), 254-264.
- Reiswig, H.M. (1981) Partial Carbon and energy budgets of the bacteriosponge Verongia fistularis (Porifera: Demospongiae) in Barbados. Marine Ecology-an Evolutionary Perspective, 2(4), 273-293.
- Santavy, D.L. and Colwell, R.R. (1990) Comparison of bacterial communities associated with the Caribbean Sclerosponge Ceratoporella nicholsoni and ambient seawater. Marine Ecology-Progress Series, 67, 73-82.
- Shanmughapriya, S., Kiran, G.S., Selvin, J., Gandhimathi, R., Baskar, T.B., Manilal, A. and Sujith, S. (2009) Optimization, production, and partial characterization of an alkalophilic amylase produced by sponge associated marine bacterium Halobacterium salinarum MMD047. Biotechnology and Bioprocess Engineering, 14(1), 67- 75.
- Karpushova, A., Brummer, F., Barth, S., Lange, S. and Schmid, R.D. (2005) Cloning, recombinant expression and biochemical characterization of novel esterases from Bacillus sp associated with the marine sponge Aplysina aerophoba. Applied Microbiology and Biotechnology, 67(1), 59-69.
- Lee, O.O., Lau, S.C.K., Tsoi, M.M.Y., Li, X.C., Plakhotnikova, I., Dobretsov, S., Wu, M.C.S., Wong, P.K., Weinbauer, M. and Qian, P.Y. (2006) Shewanella irciniae sp nov., a novel member of the family Shewanellaceae, isolated from the marine sponge Ircinia dendroides in the Bay of Villefranche, Mediterranean Sea. International Journal of Systematic and Evolutionary Microbiology, 56(12), 2871-2877.
- Hauksson, J.B., Andresson, O.S. and Asgeirsson, B. (2000) Heat-labile bacterial alkaline phosphatase from a marine Vibrio sp. Enzyme and Microbial Technology, 27(1-2), 66-73.
- Graeber, I., Kaesler, I., Borchert, M.S., Dieckmann, R., Pape, T., Lurz, R., Nielsen, P., von Dohren, H., Michaelis, W. and Szewzyk, U. (2008) Spongiibacter marinus gen. nov., sp nov., a halophilic marine bacterium isolated from the boreal sponge Haliclona sp 1. International Journal of Systematic and Evolutionary Microbiology, 58(3), 585-590.
- Zaccone, R., Caruso, G. and Cali, C. (2002) Heterotrophic bacteria in the northern Adriatic Sea: Seasonal changes and ecoenzyme profile. Marine Environmental Research, 54(1), 1-19.
- Zhang, H., Zhang, F. and Li, Z. (2009) Gene analysis, optimized production and property of marine lipase from Bacillus pumilus B106 associated with South China Sea sponge Halichodria rugosa. World Journal of Microbiology and Biotechnology, 25(7), 1267-1274.
- Mohapatra, B.R., Bapuji, M. and Sree, A. (2003) Production of industrial enzymes (Amylase, carboxymethylcellulase and protease) by bacteria isolated from marine sedentary organisms. Acta Biotechnologica, 23(1), 75-84.

