Journal of Environmental Protection
Vol. 3 No. 1 (2012) , Article ID: 16858 , 9 pages DOI:10.4236/jep.2012.31010
Hospital-Adapted Clonal Complex 17 Enterococcus faecium Found among Sand Enterococcal Isolates*
![]()
1Instituto de Biologia Experimental e Tecnológica, Oeiras, Portugal; 2Instituto de Tecnologia Química e Biológica, Oeiras, Portugal; 3Laboratorium voor Microbiologie, Vakgroep Biochemie, Fysiologie en Microbiologie, Universiteit Gent, Gent, Belgium.
Email: flopes@itqb.unl.pt
Received October 3rd, 2011; revised November 8th, 2011; accepted December 18th, 2011
Keywords: Antibiotic Resistance; Beach Sand; Clonal Complex 17; Enterococcus; Playground Sand; Virulence Factors
ABSTRACT
Though poorly studied, sand is an environment with an extended degree of interaction with man. Enterococcal strains can be found in sand but we do not know to what extent these ubiquitous opportunistic nosocomial pathogens isolated from sand carry antimicrobial resistances and virulence traits. In an attempt to fill in this knowledge gap, two distinct types of sand (beach and children playground) were examined concerning composition in enterococcal species, genetic diversity of isolates and abundance of resistance to antimicrobials and virulence traits. Five different species were found, namely Enterococcus faecium, Enterococcus faecalis, Enterococcus hirae, Enterococcus flavescens and Enterococcus casseliflavus. Although genetic diversity was evident, two different E. faecium clones, common to the two types of sand, were detected, suggesting the existence of clones well adapted to this specific environment or from a common source. E. faecium was associated with multiple antibiotic resistances, including to fluoroquinolones and tetracycline that are commonly used by veterinarians and clinicians. Among the multiresistant E. faecium strains from beach sand, two were from sequence type (ST) 442, which belongs to the wide-spread Hospital-adapted clade CC17. They both carried the esp gene and the genomic island associated with CC17. The other virulence factors screened were disseminated among E. faecalis strains, but seldom detected in the other species, evidencing the existence, in these environments, of E. faecalis strains carrying the same virulence factors as the clinical ones. The present work thus stresses the need to follow-up the presence and characterization of enterococcal strains from both beach and children playground sands and of including these environments in the epidemiological global analysis of enterococcal isolates.
1. Introduction
Enterococcus are human commensal Gram-positive bacteria, able to withstand a great diversity of environmental conditions, and probably for this reason they are commonly isolated from environments as diverse as food products [1], water and soil [2]. The presence of enterococci in these environments is probably due to fecal contamination [3]. However, the general assumption that fecal indicators, like Escherichia coli and enterococci, do not occur in natural environments such as soil or water, has recently been challenged [4], because 1) sediments could provide favorable nutrient conditions and protection from sunlight inactivation [1] and 2) enterococci could survive desiccation and regrow in rewetted sediments [5]. In fact, in the last years a few authors reported the presence of enterococci in sand [3-6].
For many years members of the genus Enterococcus were considered harmless, but this view has changed. Nowadays they are also seen as human nosocomial pathogens, emerging as the second most frequently reported cause of surgical wound and nosocomial urinary tract infections and the third most frequently reported cause of bacteraemia [1]. The emergence of enterococci as nosocomial infectious agents is related to the use of antibiotics and to the fact that these bacteria are intrinsically resistant to many of the antibiotics used in clinical settings. Recently, enterococcal isolates from different sources have been screened for resistance to some of the more clinically important antibiotics [7-10], and results have shown that resistance is present in almost all environments, although to different extents. This recent awareness, and consequent concern, of enterococci as a health problem has led researchers to invest a lot of efforts to understand the factors that influence the relationship of these organisms with their host, i.e. the virulence factors. Several virulence factors have been identified, including aggregation substance, surface protein Esp and adhesin Ace, all playing a role in adhesion to host cells and tissues; cytolysin, gelatinase and hyaluronidase, which are responsible for tissue damage [11]; bile salt hydrolase, contributing to survival in the gastrointestinal tract [12]; and the Enterococcus faecalis endocarditis antigen (EfaA), for which the role in virulence has not yet been completely clarified [13]. All these factors have been studied in E. faecalis, although some of them have been also reported in other species of the genus [14] and in strains not associated with nosocomial environments [15].
The presence of enterococci in sand, though acknowledged, has not been subject of a more thorough analysis that could provide information on how this environment can contribute to the general trafficking of antibiotic resistance and virulence determinants carried by enterococci. Thus, the objectives of the present study were to determine the genetic diversity, antibiotic resistance and carriage of virulence determinants by enterococci resident in sand in Portugal. Beach sand has been sampled by others. Therefore, we included in this study a sample of children playground sand, which, to our knowledge, has never been studied as an environmental source of enterococci.
2. Materials and Methods
2.1. Microorganisms
A total of 97 enterococal isolates from beach (78 isolates) and playground sand (19 isolates) were collected and sent to our Laboratory after primary identification to the genus level. In the Lab, all microorganisms were grown in Brain Hearth Infusion (BHI) (Oxoid, Hampshire, UK) at 37˚C, unless otherwise mentioned. For identification purposes Enterococcus type-strains obtained from the Deutsch Sammlung von Mikroorganismen and Zellkulturen (DSMZ; Braunschweig, Germany), were used as references: Enterococcus casseliflavus DSM 20680, Enterococcus dispar DSM 6630, Enterococcus durans DSM 20633, Enterococcus faecalis DSM 20478, Enterococcus faecium DSM 20477, Enterococcus flavescens DSM 7370, Enterococcus gallinarum DSM 20628, Enterococcus hirae DSM 20160, Enterococcus mundtii DSM 4838, Enterococcus raffinosus DSM 5633, Enterococcus solitarius DSM 5634. Enterococcus faecalis DSM 2570 was used as the control strain in the disk antimicrobial susceptibility assays.
2.2. DNA Preparation
Total DNA extraction was performed as described before [16] with minor changes: cells were harvested and resuspended in 3/20 of the initial volume of TES and incubated in the presence of lysozyme (5 mg/mL) (Sigma, Steinheim, Germany) for 40 minutes. 300 µL of saline solution and 40 µL of SDS 20% (w/v) (Sigma) were added and mixed by inversion. Phenol extractions and ethanol precipitation were preformed and the final product was treated with RNase (10 µg/mL) (Sigma).
2.3. Identification Procedures
All isolates were screened by PCR with species specific primers (Table 1). Since E. faecalis and E. faecium are the most abundant enterococcal species, all 97 isolates were screened using primers for both species. Those not identified as one of these two species, were then tested, sequentially and using the same approach, with primers for E. durans, E. hirae, E. casseliflavus, E. mundtii, E. dispar, E. flavescens, E. gallinarum, E. raffinosus and E. solitarius, as described in Table 1. After this procedure 41 isolates remained unidentified. All isolates were typed using PFGE and among these 41 unidentified isolates we

Table 1. Primers used to amplify specific genes from each species.
detected 18 types. One isolate from each type was selected for repetitive sequence-based PCR fingerprinting with the (GTG)5 primer [17]. Fingerprints were analyzed with BioNumerics version 3.0 software (Applied Maths, Sint Martens Latem, Belgium) using the Pearson Correlation Coefficient and UPGMA for pattern analysis, and were compared with available data for enterococcal reference strains [17]. For strains which were not identified by the (GTG)5-PCR approach, part of the pheS gene was amplified and sequenced, as described before [18]. Sequences were analyzed by using the BioNumerics version 3.0 software and compared with sequences of enterococcal reference strains present in public databases. The remaining unidentified isolates (23 isolates) which were not selected for (GTG)5-PCR and pheS analyses were genetically indistinguishable, as determined by PF GE, from the ones that were analyzed and were therefore assumed to represent the same species.
2.4. Pulsed Field Gel Electrophoresis
PFGE was performed as described before [19].
2.5. Antimicrobial Susceptibility Test
The susceptibility of enterococcal strains to antibiotics was determined using the disk diffusion method according to CLSI [20]. Antibiotics tested and disk content were as described before [1]. All isolates were cultured overnight in Mueller-Hinton Broth (MHB) (Oxoid, Hampshire, UK), with the exception of four isolates which presented impaired growth in this medium and for this reason were grown in BHI broth. MIC was determined, for a few isolates and antibiotics, using E-test from AB Biodisk (Solna, Sweden) according to manufacturer instructions.
2.6. Screening of Virulence Factors
Gelatinase activity was verified as described before [14]. Blood agar plates (BioMerieux, Marcy l’Etoile, France) were used to detect hemolytic activity and, after inoculation, plates were incubated for 48 hours in anaerobic conditions before assessment of that activity. All isolates were tested for the presence of fsrA, fsrB, fsrC, gelE, sprE, ace, efaAfs and asa1 genes. PCR amplifications were performed in a T-personal Combi thermocycler using primers targeting these genes [21]. Screening for virulence factors Hyl and Esp was performed using the following primers and only for a few E. faecium isolates, as described ahead:
hylEfmf (5’-GTTAGAAGAAGTCTGGAAACCG-3’), hylEfmr (5’-TGCTAAGATATTCCTCTACTCG-3’), espEfmf (5’-TGCTAATGCTAGTCCACGACC-3’) and espEfmr (5’-GCGTCAACACT TGCATT GCCGA-3’) [22]. Reference [23] identified a new genomic Island (GI) specific to hospital-acquired strains belonging to CC17. This genomic island was composed by 7 genes (orf1474 to orf1483) encoding a potential new metabolic pathway involved in the metabolism and transport of carbohydrates. To determine the presence of this GI in some of our E. faecium isolates, orf1477 was amplified by PCR using the following set of primers: 1477F (5’-CATTACTGTATTGGGCTTCGA-3’) and 1477R (5’-CTCTATGGTATGCTTCTGCTCC-3’).
2.7. Multilocus Sequence Typing (MLST)
Five unrelated (by PFGE) E. faecium isolates, resistant to more then 20 antibiotics by disk diffusion method, were selected for MLST typing. Internal fragments of seven housekeeping genes were amplified by PCR with the sets of primers described before [24]. Sequencing was done at Baseclear (Leiden, Netherlands). MLST alleles and sequence types (ST) were identified using the database (http://efaecium.mlst.net/).
3. Results and Discussion
Beaches are quite dynamic ecosystems, subject to influence from man, land, wind and rain and sea water. Playground sand is a different ecosystem, influenced by humans, land, rain and wind. Together, they constitute important reservoirs of microorganisms and are vehicles for human cross-contamination. Enterococci are opportunistic human nosocomial pathogens, with a recognized ability to survive outside environments, such as food, soil and water. The omnipresence of these bacteria in and out of the human host, together with their ability to exchange genetic material, allows them to play an important role in transmission, between environments, of both strains and genes coding for antibiotic resistance and virulence determinants. Although acknowledged as a site of contamination with enterococci [2-6,25,26], sand has not been very well explored and characterized as a reservoir of Enterococcus strains. The presence of enterococci in sands has been pointed [6] as a possible cause of water quality failures. Finding enterococci in water and sand is relevant because they can be a vehicle for infection and/or carriage of potentially virulent strains and eventually contribute to the increasing number of infections caused by these bacteria. In order to understand the role of these environments in the global epidemiology of enterococcal strains we must, first, characterize the genetic diversity of the collected isolates and also two most important factors relevant for infection, namely carriage of antibiotic resistance and virulence.
The isolates of the present study were collected from a children playground in the Lisbon area (19 isolates) and from sand from the Sesimbra beach (78 isolates). The latter is an Atlantic beach, located approximately 30 km away from the nearest hospital and there is no sewage or waste water being deposited near the beach. As in other reports [6,9,10], the predominant species found were E. faecium (46%) and E. faecalis (33%), but other species were also detected, including E. hirae (8%), E. flavescens (5%) and E. casseliflavus (5%). E. mundtii, E. durans, E. avium and E. gallinarum were not detected although they have also been associated with sand [2,6]. However, the distribution of species was different between samples: in the playground sand we found 42% of E. faecium and only 5% of E. faecalis, whereas in beach sand the frequency of the same species was 50% and 41%, respectively, and E. flavescens was not detected. No obvious reason or deduction can be withdrawn from these data, but it is clear that E. faecalis is less represented in the playground sample.
Relevant findings from the antibiotic resistance screening are summarized in Table 2. We included in this study antibiotics for which the genus Enterococcus is considered intrinsically resistant or susceptible because it would be possible, in sand, to find different behavior, as we did previously in food isolates [27]. Analysis of Table 2 shows an association between E. faecium and resistance to fluoroquinolones, tetracyclines and b-lactams. For all other antibiotics tested, the results obtained revealed a similar behavior of E. faecium and E. faecalis isolates. Overall, the enterococcal isolates studied were resistant to bacitracin (90%), colistin (97%), kanamycin (83%), lincomycin (80%), methicillin (97%), polymixin B (97%) and susceptible to amoxicillin, ampicillin, chloramphenicol (97%), imipenem, penicillin, piperacillin, vancomycin (90%) and sulphamethoxazole/trimethopim (96%). These results are similar to data previously reported for environmental enterococcal strains from food, and also corroborate resistance and susceptibilities common to the genus Enterococcus [1]. Resistance to norfloxacin, ciprofloxacin, enrofloxacin, erythromycin, nitrofurantoin and vancomycin observed in sand enterococci
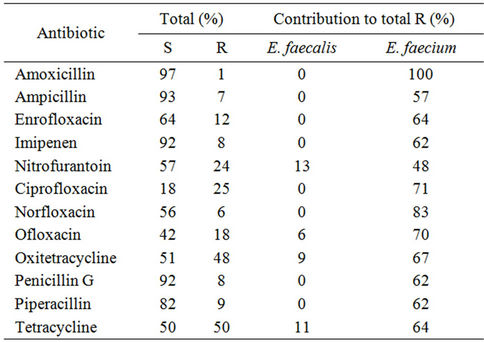
Table 2. Percentage of isolates which were found to be resistant (R) and susceptible (S) to antibiotics showing different behaviors in the two most representative species.
is at the same level as reported in enterococci from other non clinical environments such as dairy products [1,8], wastewater [7], animal food products and animals [9]. Concerning tetracyclin and bacitracin we observed resistances similar to the ones found in clinical enterococcal isolates (50% for tetracycline and 90% for bacitracin). The enterococci studied showed higher resistances to kanamycin and rifampicin than the enterococci isolated from wild animals [10] and wastewater [8]. Altogether, these observations point out that we can find in sand more resistant isolates than in other non-clinical environments and raises the question about the primary origin of these isolates.
All 97 isolates were typed using PFGE, band patterns were analyzed using the Tenover criteria [28], and isolates were considered genetically related accordingly. A total of 46 PFGE types were defined, assuming that related isolates have similarity higher than 96% (Figure 1). In our study we found a diversity of 2.6 and 1.8 for E. faecalis and E. faecium, respectively. Diversity was calculated by dividing the number of strains of one species by the number of PFGE types for that species. Overall, these results demonstrate a high genetic diversity which appears to be a common feature in the genus Enterococcus as it has been described in other environments as well. Although most of the PFGE types were composed of isolates from the same sand type, we found two PFGE types, among E. faecium species, with isolates from both beach and playground sand. This suggests the existence of E. faecium clones well adapted to this specific environment, sand. This result is quite interesting, because the two sand samples are not only spatially distant but also subject to different environmental influences and contamination sources. The fact that we found the same clones in these two samples points out that not only sand can be a reservoir of enterococcal strains, but also that these environments should start to be included in the epidemiological global analysis of enterococcal isolates.
One of the E. faecium clones common to the two sand samples was found to be resistant to 23 antibiotics, among the 30 tested. Three other E. faecium isolates (two from beach and one from playground) and two E. hirae isolates from playground sand, also showed resistance to more than 20 antibiotics (Figure 1). Resistance to fluoroquinolones has been, in the last years, found to be associated with E. faecium nosocomial isolates. It is interesting, but also a matter of concern, that E. faecium isolated from sand, both from a beach located away from hospitals, and from a children playground, are also, as the nosocomial strains, associated with the same antimicrobial resistances. E. faecalis is the species more frequently associated with nosocomial infection, accounting for up to 80% of all enterococcal nosocomial infections [29]. However, E. faecium is associated with multidrug
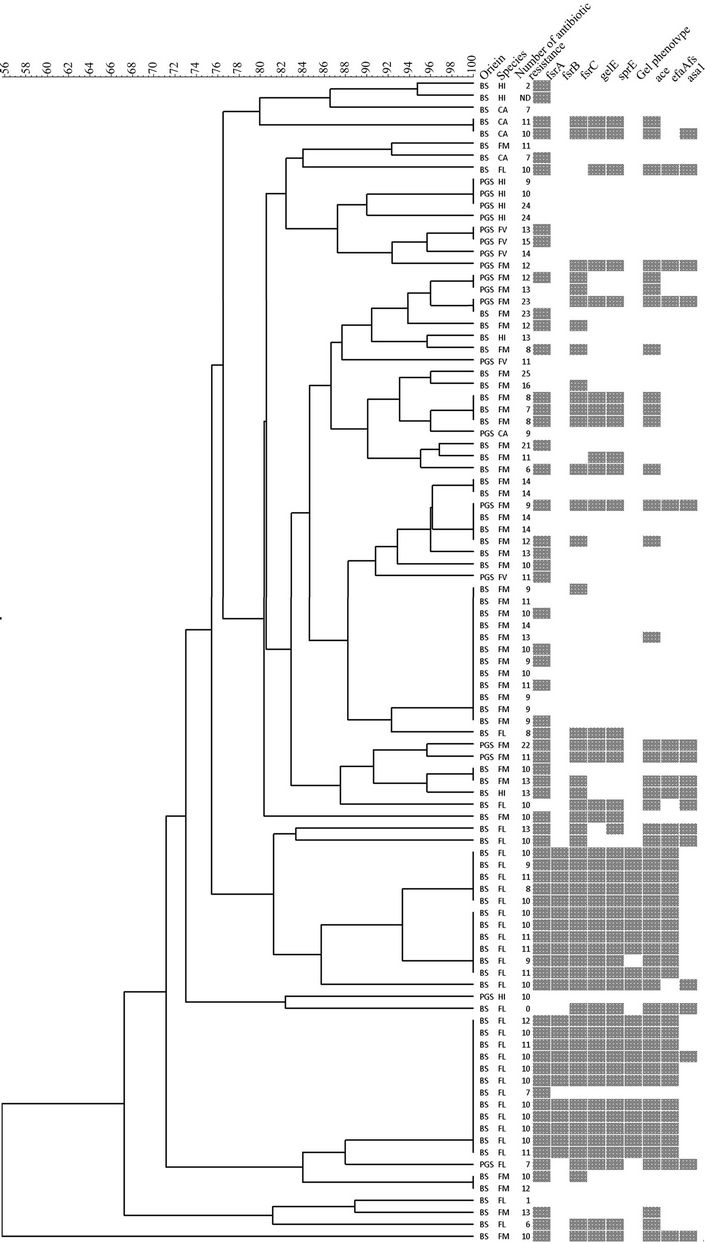

Figure 1. Dendrogram representing all studied isolates grouped according to PFGE type.
resistance. This species has evolved in the last 15 years from an avirulent commensal to the third most frequently isolated nosocomial pathogen among intensive care unit patients in the United States [30]. Molecular epidemiological studies of E. faecium using MLST revealed the existence of a distinct genetic subpopulation, named clonal complex 17 (CC17), responsible for the majority of hospital-related infections and outbreaks. CC17 has spread globally [31] and seems well adapted to the Hospital settings as well as associated with high-level ciprofloxacin resistance and ampicillin resistance [28]. It appears that the acquisition of ampicilin resistance was one of the first steps in the adaption of E. faecium to hospital environment facilitating the subsequent emergence of vancomycin resistance [32]. We thus decided to investigate further the five E. faecium strains resistant to more then 20 antibiotics and type them by MLST. Two of the strains (16D7 and S1-2), constituting one of the clones found simultaneously in PGS and BS, although confirmed as E. faecium by both ddl and sodA specific PCR, did not amplify any of the seven genes of the MLST scheme. It was thus impossible to type these two isolates by MLST. This typing method has been developed with E. faecium isolates from similar environments, namely human and animals, both commensals and from clinical infections, as already mentioned. We thus reinforce the possibility that environmental strains, such as the ones we isolated from sand, have differences in the sequences of the house-keeping genes used for the MLST scheme. These isolates originate from an unusual source that has not been sampled before, which could suggest the existence of a distinct E. faecium subpopulation with deviant MLST genes. For the other three isolates, MLST was carried out successfully. Two of them (5LM6 and 5LM8, both from beach sand) were ascribed to the new ST442 (atpA 5, ddl 1, gdh 1, purK 2, gyd 6, pstS 1 and adk 1) and one (14D5, from playground sand) to the new ST470 (atpA 25, ddl 13, gdh 34, purK 48, gyd 19, pstS 26 and adk 6). When eBURST analysis, comparing the entire MLST database, was done it was clear that ST442 is still part of the CC17 (Figure 2). ST442 is a single-locus variant (SLV) of ST324 and a double-locus variant (DLV) of six other ST’s: ST416, ST121, ST55, ST323 (co-founder), ST92 (co-founder) and ST313. CC17 is also characterized
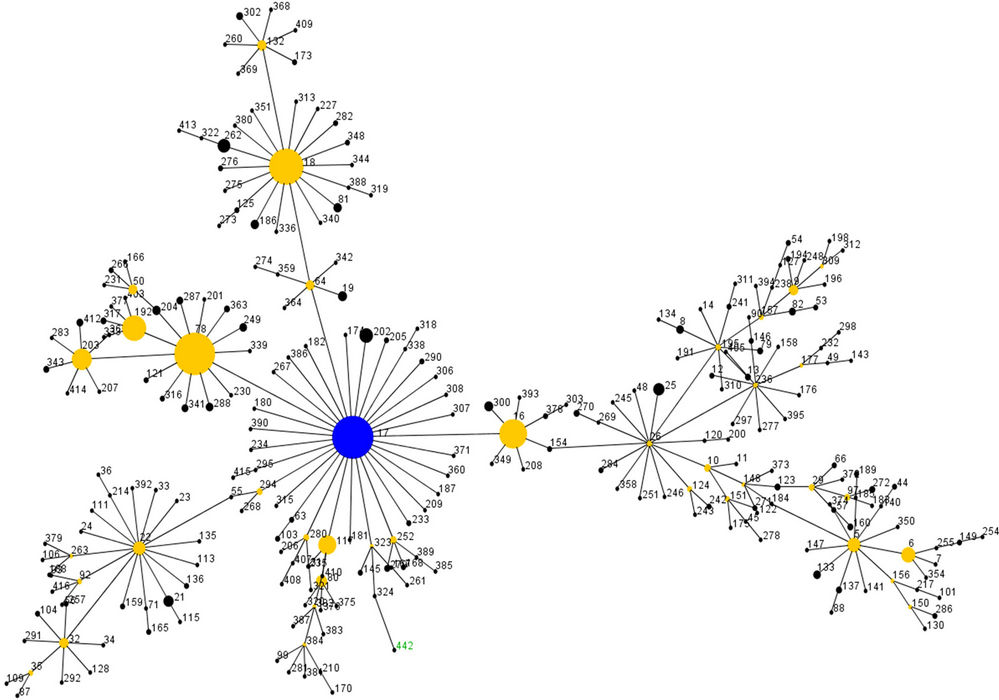

Figure 2. eBURST analysis of CC17 strains.
by the presence of a GI, which includes the esp gene. We detected both esp and orf1477 in the CC17 sand strains but hyl gene was absent in the same strains. These two CC17 sand strains were vancomycin and ampicillin sensitive (MICs of 2 mg/mL and 1 mg/mL, respectively). One, 5LM6, was confirmed as resistant to fluoroquinolones by MIC determination (>32 mg/mL for ciprofloxacin, ofloxacin and enrofloxacin and 16 mg/mL for norfloxacin). None of these two strains carried any of the virulence factors screened other then those already mentioned, namely esp and the PAI. Our results clearly show that CC17, which until this moment includes mainly hospital strains and some strains isolated from calf, pigs and dogs, is also disseminating into “abiotic” environments, like beach sand, where the selective pressure of antibiotics is most likely irrelevant. It is possible that these CC17 sand strains, bearing some of the characteristics of the CC17 nosocomial strains, were brought to that environment by man or animals, most likely pets (dogs). Further studies need to be carried out to understand this phenomenon. Our findings stress, and urge, the need to closely survey and include sand, and eventually other “abiotic” environments influenced by man, in the epidemiological evaluation of enterococcal strain trafficking.
Research on enterococcal virulence has demonstrated that factors, initially ascribed a role in E. faecalis virulence, are in fact disseminated in the genus and are found in non-clinical environments [15]. Analysis of Figure 1 clearly shows that the virulence factors screened in this study are disseminated among E. faecalis, evidencing the existence in sand of E. faecalis strains carrying the same virulence factors as the clinical ones. This fact, however, does not imply any correlation between E. faecalis sand strains and pathogenicity. The same virulence traits were seldom detected in the other species, somehow contradicting the previous assumption that E. faecalis virulence determinants are common in the genus Enterococcus.
The screening of ace, efaAfs and asa1 genes revealed that these genes were absent in 48% of the isolates, 52% were positive to ace gene, 34% were positive to efaAfs and 16% to asa1. E. faecalis genes coding for surface proteins were found in other species, namely E. hirae, E. faecium and E. casseliflavus. Until now, to our knowledge, these virulence factors were only reported in E. faecalis and E. faecium. None of the strains studied was hemolytic and 23% produced gelatinase. All gelatinase producers were E. faecalis (Figure 1). The screening of genes involved in gelatinase expression (fsrA, fsrB, fsrC and gelE) showed that all the isolates with gelatinase activity were also positive for all the genes screened, as expected from previous work [14]. We were able to detect all genes screened in one of the isolates without gelatinase activity. This silent behavior of the gelatinase operon has been reported previously [14], but reasons for the discrepancy between genotype and phenotype have not yet been reported. Gelatinase activity is associated with organisms that are able to cause infection. However, there is also one report of this virulence factor in food associated enterococci [14] and in this work we were able to detect gelE gene, the fsr operon and also the gelatinase phenotype in environmental isolates, presumably not associated with human infections. The presence of virulence factors in the sand enteroccal isolates does not preclude, per se, the pathogenic potential of the same bacteria. However, it reveals that enterococci colonizing, or simply surviving, in “abiotic” environments carry the same genes which, in the nococomial environment, have proven relevant for the infection induced by E. faecalis. We cannot exclude the possibility that the virulence factors we found in the sand isolates are relevant for the survival and establishment of these bacteria in the sand.
In summary, PFGE revealed the presence of two different E. faecium clones in both sand samples, suggesting the existence of clones well adapted to this specific environment. E. faecium was associated with multiple antimicrobial resistances (five strains were resistant to more then 20 antibiotics) and in particular to fluoroquinolones and tetracycline. The virulence factors screened were disseminated among E. faecalis strains, but seldom detected in the other species, evidencing the existence, in these environments, of E. faecalis strains carrying the same virulence factors as the clinical ones. Finally, we detected two E. faecium strains belonging to the hospital clade CC17, carrying the esp gene, the GI and resistance to fluoroquinolones. Enterococci are able to colonize environments traditionally not colonized by fecal bacteria. These bacteria are resistant to salt and have been found before in sand. However, the high frequencies of resistance to some antibiotics were unexpected as was the resemblance of some strains to hospital-adapted strains. These results strongly advise monitoring beach and playground sands, places where the contact between humans and sediments is important and highlights the imperative need to include sand in the epidemiological global analysis of enterococcal isolates, together with its characterization concerning antibiotic resistance and presence of virulence traits.
4. Acknowledgements
The authors acknowledge Professor Alexandra Nogueira Silva, from Faculdade de Farmácia de Lisboa, for giving us the enterococcal isolates, and Teresa Braga for confirmation of some results.
REFERENCES
- M. F. S. Lopes, T. Ribeiro, M. Abrantes, J. J. F. Marques, R. Tenreiro and M. T. B. Crespo, “Antimicrobial Resistance Profiles of Dairy and Clinical Isolates and Type Strains of Enterococci,” International Journal of Food Microbiology, Vol. 103, No. 2, 2005, pp. 191-198. doi:10.1016/j.ijfoodmicro.2004.12.025
- A. J. de Oliveira and J. M. Watanabe-Pinhata, “Antimicrobial Resistance and Species Composition of Enterococcus spp. Isolated from Waters and Sands of Marine Recreational Beaches in Southeastern Brazil,” Water Research, Vol. 42, No. 8-9, 2008, pp. 2242-2250. doi:10.1016/j.watres.2007.12.002
- E. W. Alm, J. Burke and A. Spain, “Fecal Indicator Bacteria Are Abundant in Wet Sand at Freshwater Beaches,” Water Research, Vol. 37, No. 16, 2003, pp 3978-3982. doi:10.1016/S0043-1354(03)00301-4
- R. L. Whitman, D. A. Shively, H. Pawlik, M. B. Nevers and M.N. Byappanahalli, “Occurrence of Escherichia coli and Enterococci in Cladophora (Chlorophyta) in Nearshore Water and Beach Sand of Lake Michigan,” Applied and Environmental Microbiology, Vol. 69, No. 8, 2003, pp. 4714-4719. doi:10.1128/AEM.69.8.4714-4719.2003
- P. G. Hartel, K. Rodgers, J. A. Fisher, J. L. McDonald, L. C. Gentit, E. Otero, Y. Rivera-Torres, T. L. Bryant and S. H. Jones, “Survival and Regrowth of Fecal Enterococci in Desiccated and Rewetted Sediments,” Proceedings of the 2005 Georgia Water Resources Conference, Athens, 25-27 April 2005.
- D. M. Ferguson, D. F. Moore, M. A. Getrich and M. H. Zhowandai, “Enumeration and Speciation of Enterococci Found in Marine and Intertidal Sediments and Costal Water in Southern California,” Journal of Applied Microbiology, Vol. 99, No. 3, 2005, pp. 598-608. doi:10.1111/j.1365-2672.2005.02660.x
- P. M. M. da Costa, P. M. Vaz-Pires and F. M. Bernardo, “Antibiotic Resistance of Enterococcus spp. Isolated from Wastewater and Sludge of Poultry Slaughterhouses,” Journal of Environmental Science and Health B, Vol. 41, No. 8, 2006, pp. 1393-1403. doi:10.1080/03601230600964258
- L. Mannu, A. Paba, E. Daga, R. Comunian, S. Zanetti, I. Duprè and L. A. Sechi, “Comparision of the Incidence of Virulence Determinants and Antibiotic Resistance between Enterococcus faecium Strains of Dairy, Animal and Clinical Origin,” International Journal of Food Microbiology, Vol. 88, No. 2-3, 2003, pp. 291-304. doi:10.1016/S0168-1605(03)00191-0
- J. Peters, K. Mac, H. Wichmann-Schauer, G. Klein and L. Ellerbroek, “Species Distribution and Antibiotic Resistance Patterns of Enterococci Isolated from Food of Animal Origin in Germany,” International Journal of Food Microbiology, Vol. 88, No. 2-3, 2003, pp. 311-314. doi:10.1016/S0168-1605(03)00193-4
- P. Poeta, D. Costa, Y. Sáenz, N. Klibi, F. Ruiz-Larrea and C. Torres, “Characterization of Antibiotic Resistance Genes and Virulence Factors in Faecal Enterococci of Wild Animals in Portugal,” Journal of Veterinary Medicine. B Infectious Disease and Veterinary Public Health, Vol. 52, No. 9, 2005, pp. 396-402.
- G. Kayaoglu and D. Orstavik, “Virulence Factors of Enterococcus faecalis: Relationship to Endodontic Disease,” Critical Reviews in Oral Biology and Medicine, Vol. 15, No. 5, 2004, pp. 308-320. doi:10.1177/154411130401500506
- S. M. McBride, V. A. Fischetti, D. J. Leblanc, R. C. Moellering Jr. and M. S. Gilmore, “Genetic Diversity among Enterococcus faecalis,” PLoS ONE, Vol. 2, No. 7, 2007, p. e582. doi:10.1371/journal.pone.0000582
- A. M. Lowe, P. A. Lambert and A. W. Smith, “Cloning of an Enterococcus faecalis Endocarditis Antigen: Homology with Adhesins from Some Oral Streptococci,” Infection and Immunity, Vol. 63, No. 12, 1995, pp. 703-706.
- M. F. S. Lopes, A. P. Simões, R. Tenreiro, J. J. Marques and M. T. Crespo, “Activity and expression of a Virulence Factor, Gelatinase, in Dairy Enterococci,” International Journal of Food Microbiology, Vol. 112, No. 3, 2006, pp. 208-214. doi:10.1016/j.ijfoodmicro.2006.09.004
- T. Semedo, M. A. Santos, M. F. Lopes, J. J. Figueiredo Marques, M. T. Barreto Crespo and R. Tenreiro, “Virulence Factors in Food, Clinical and Reference Enterococci: A Common Trait in the Genus?” Systematic and Applied Microbiology, Vol. 26, No. 1, 2003, pp. 13-22. doi:10.1078/072320203322337263
- P. Serror, T. Sasaki, S. D. Ehrlich and E. Maguin, “Electrotransformation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis with Various Plasmids,” Applied and Environmental Microbiology, Vol. 68, No. 1, 2002, pp. 46-52. doi:10.1128/AEM.68.1.46-52.2002
- P. Svec, M. Vancanneyt, M. Seman, C. Snauwaert, K. Lefebvre, I. Sedlácek and J. Swings, “Evaluation of (GTG)5- PCR for Identification of Enterococcus spp.,” FEMS Microbiology Letters, Vol. 247, No. 1, 2005, pp. 59-63. doi:10.1016/j.femsle.2005.04.030
- S. M. Naser, F. L. Thompson, B. Hoste, D. Gevers, P. Dawyndt, M. Vancanneyt and J. Swings, “Application of Multilocus Sequence Analysis (MLSA) for Rapid Identification of Enterococcus species Based on rpoA and pheS Genes,” Microbiology, Vol. 151, No. 7, 2005, pp. 2141- 2150. doi:10.1099/mic.0.27840-0
- T. Ribeiro, M. Abrantes, M. F. Lopes and M. T. Crespo, “Vancomycin-Susceptible Dairy and Clinical Enterococcal Isolates Carry vanA and vanB Genes,” International Journal of Food Microbiology, Vol. 113, No. 3, 2007, pp. 289-295. doi:10.1016/j.ijfoodmicro.2006.08.010
- National Committee for Clinical Laboratory Standards, Performance Standards for Antimicrobial Susceptibility Testing, Eleventh-Informational Supplement. Disk diffusion, M100-S11, NCCLS, Villanova, 2001,.
- T. C. Ribeiro, V. Pinto, F. Gaspar, M. F. S. Lopes, “Enterococcus hirae Causing Wound Infections in a Hospital,” Journal of Clinical Chinese Medicine, Vol. 3, No. 3, 2008, pp. 150-152. doi:10.1086/367711
- L. B. Rice, L. Carias, S. Rudin, C. Vael, H. Goossens, C. Konstabel, I. Klare, S. R. Nallapareddy, W. Huang and B. E. Murray, “A Potential Virulence Gene, hylEfm, Predominates in Enterococcus faecium of Clinical Origin,” Journal of Infectious Disease, Vol. 187, No. 3, 2003, pp. 508-512.
- E. Heikens, W. van Schaik, H. L. Leavis, M. J. M. Bonten and R. J. L. Willems, “Identification of a Novel Genomic Island Specific to Hospital-Acquired Clonal Complex 17 Enterococcus faecium Isolates,” Applied and Environmental Microbiology, Vol. 74, No. 22, 2008, pp. 7094- 7097. doi:10.1128/AEM.01378-08
- W. L. Homan, D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. A. van Embden and R. J. L. Willems, “Multilocus Sequence Typing Scheme for Enterococcus faecium,” Journal of Clinical Microbiology, Vol. 40, No. 6, 2002, pp. 1963-1971. doi:10.1128/JCM.40.6.1963-1971.2002
- H. Li-Ming and H. Zhenli, “Water Quality Prediction of Marine Recreational Beaches Receiving Watershed Baseflow and Stormwater Runoff in Southern California, USA,” Water Research, Vol. 42, No. 10-11, 2008, pp. 2563-2573. doi:10.1016/j.watres.2008.01.002
- A. Pianetti, F. Bruscolini, L. Sabatini and P. Colantoni, “Microbial Characteristics of Marine Sediments in Bathing Area along Pesaro-Gabicce Coast (Italy): A Preliminary Study,” Journal of Applied Microbiology, Vol. 97, No. 4, 2004, pp. 682-689. doi:10.1111/j.1365-2672.2004.02352.x
- H. L. Leavis, R. J. Willems, W. J. van Wamel, F. H. Schuren, M. P. Caspers and M. J. Bonten, “Insertion Sequence-Driven Diversification Creates a Globally Dispersed Emerging Multiresistant Subspecies of E. faecium,” PLoS Pathogens, Vol. 3, No. 1, 2007, p. e7. doi:10.1371/journal.ppat.0030007
- H. L. Leavis, M. J. Bonten and R. J. Willems, “Identification of High-Risk Enterococcal Clonal Complexes: Global Dispersion and Antibiotic Resistance,” Current Opinion in Microbiology, Vol. 9, No. 5, 2006, pp. 454-460. doi:10.1016/j.mib.2006.07.001
- M. Kawalec, J. Kedzierska, A. Gajda, E. Sadowy, J. Wegrzyn, S. Naser, A. B. Skotnicki, M. Gniadkowski and W. Hryniewicz, “Hospital Outbreak of Vancomycin-Resistant Enterococci Caused by a Single Clone of Enterococcus raffinosus and Several Clones of Enterococcus faecium,” Clinical Microbiology Infections, Vol. 13, No. 9, 2007, pp. 893-901. doi:10.1111/j.1469-0691.2007.01774.x
- J. Top, R. Willems, H. Block, M. de Regt, K. Jalink, A. Troelstra, B. Goorhuis and M. Bonten, “Ecological Replacement of Enterococcus faecalis by Multiresistant Clonal Complex 17 Enterococcus faecium,” Clinical Microbiology of Infections, Vol. 13, No. 3, 2007, pp. 316- 319. doi:10.1111/j.1469-0691.2006.01631.x
- M. F. Lopes, T. Ribeiro, M. P. Martins, R. Tenreiro and M. T. Crespo, “Gentamicin Resistance in Dairy and Clinical Enterococcal Isolates and in Reference Strains,” Journal of Antimicrobial Chemotherapy, Vol. 52, No. 2, 2003, pp. 214-219. doi:10.1093/jac/dkg304
- F. C. Tenover, R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persin and B. Swaminathan, “Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed-Field Gel Electrophoresis: Criteria for Bacterial Strain Typing,” Journal of Clinical Microbiology, Vol. 33, No. 9, 1995, pp. 2233-2239.
- C. R. Jackson, P. J. Fedorka-Cray and J. B. Barrett, “Use of a GenusAnd Species-Specific Multiplex PCR for Identification of Enterococci,” Journal of Clinical Microbiology, Vol. 42, No. 8, 2004, pp. 3558-3565. doi:10.1128/JCM.42.8.3558-3565.2004
- C. A. Arias, B. Robredo, K. V. Singh, C. Torres, D. Panesso and B. E. Murray, “Rapid Identification of Enterococcus hirae and Enterococcus durans by PCR and detection of a Homologue of the E. hirae mur-2 Gene in E. durans,” Journal of Clinical Microbiology, Vol. 44, No. 8, 2006, pp. 1567-1570. doi:10.1128/JCM.44.4.1567-1570.2006
- F. Depardieu, B. Perichon and P. Courvalin, “Detection of the Van Alphabet and Identification of Enterococci and Staphylococci at the Species Level by Multiplex PCR,” Journal of Clinical Microbiology, Vol. 42, No. 12, 2004, pp. 5857-5860. doi:10.1128/JCM.42.12.5857-5860.2004
NOTES
*The authors acknowledge financial support from the Fund for Scientific Research, Flanders, Fundação para a Ciência e Tecnologia, Portugal, through project grant POCTI/BIA-BCM/60643/2004 co-financed through FEDER, and grant PEst-OE/EQB/LA0004/2011.

