American Journal of Plant Sciences
Vol.3 No.3(2012), Article ID:17921,8 pages DOI:10.4236/ajps.2012.33045
Evolutionary Relationship of Wheat Protein Disulphide Isomerase (PDI) Gene Promoter Sequence Based on Phylogenetic Analysis
![]()
1Plant Genetic Resources, Scuola Superiore Sant’ Anna, Rome, Italy; 2Division of Plant Sciences, University of Missouri, Columbia, USA.
Email: a.dhanapal@sssup.it, dhanapala@missouri.edu
Received December 12th, 2011; revised January 10th, 2012; accepted January 24th, 2012
Keywords: Protein Disulfide Isomerase (PDI) promoter; wheat; phylogenetic analysis
ABSTRACT
Protein disulphide isomerase (PDI) is an oxidoreductase enzyme abundant in the endoplasmic reticulum (ER). In plants, PDIs have been shown to assist the folding and deposition of seed storage proteins during the biogenesis of protein bodies in the endosperm. Cloning and characterization of the complete set of genes encoding PDI and PDI like proteins in bread wheat (Triticum aestivum cv. Chinese Spring) and the comparison of their sequence, structure and expression with homologous genes from other plant species were reported in our previous publications. Promoter sequences of three homoeologous genes encoding typical PDI, located on chromosome group 4 of bread wheat, and PDI promoter sequence analysis of Triticum urartu, Aegilops speltoides and Aegilops tauschii had also been reported previously. In this study, we report the isolation and sequencing of a ~700 bp region, comprising ~600 bp of the putative promoter region and 88 bp of the first exon of the typical PDI gene, in five accessions each from Triticum urartu (AA), Aegilops speltoides (BB) and Aegilops tauschii (DD). Sequence analysis indicated large variation among sequences belonging to the different genomes, while close similarity was found within each species and with the corresponding homoeologous PDI sequences of Triticum aestivum cv. CS (AABBDD) resulting in an overall high conservation of the sequence conferring endosperm-specific expression.
1. Introduction
Wheat is adapted to temperate regions of the world and was one of the first crops to be domesticated. Spread over all continents, it is today one of the most important food source for human beings. Bread wheat belongs to the tribe Triticeae, and has three genomes A, B and D, each organised in seven homoeologous chromosome groups. The diploid progenitors of the three genomes A, B, and D have been identified in Triticum urartu, Aegilops speltoides and Aegilops tauschii, although the progenitor of B genome is still matter of debate [1]. Throughout their evolutionary history, multiple polyploidization events occurred between species of the Triticum and Aegilops genera and human manipulation of wild species led to the domestication of different cultivated lineages [2-5]. The first hybridisation event combined the genome of the wild species Triticum urartu (AA genome) and that of Aegilops speltoides (BB genome) to give rise to the tetraploid (AABB) Triticum turgidum ssp. dicoccoides. Subsequent molecular studies identified Triticum urartu as the donor of the A genome of emmer wheat (AABB), durum wheat (AABB), and common wheat (AABBDD) [6-10]. The second polyploidisation event occurred between the new tetraploid species (AABB) and diploid Aegilops tauschii (DD genome) to give rise to hexaploid wheat (AABBDD) Triticum aestivum [8,11]. A recent study revealed lower levels of polymorphism in the D genome than in the A and B genome, indicating that possibly the gene flow from tetraploid to hexaploid species was more frequent than that from the diploid Aegilops tauschii [12].
Wheat grains are consumed under many different forms, such as flour for leavened, flat and steamed breads, biscuits, cookies, cakes, breakfast cereal, pasta, noodles, couscous and for fermentation to make beer, alcohol, vodka and biofuel. It is composed by starch, proteins and other compounds, which accumulate in significant quantities during its development, in particular grain storage proteins, are responsible for the quality of the end product. The majority of them are prolamins, which account for >90% of the total protein content in the wheat grain. Different quality of grain storage proteins produced by the same genotype under different environmental condition has stimulated research on protein folding and assembling.
Secretory protein folding and disulphide bond formation takes place within the Endoplasmic reticulum (ER lumen), but the precise mechanisms involved, and the role of other proteins such as molecular chaperones, are not fully understood [13]. PDI plays an important role in assisting protein folding and assembly, catalyzing thioldisulfide oxidation, reduction and isomeration, this latter occurring directly by intramolecular disulfide rearrangement or through cycles of reduction and oxidation [14]. During the maturation of the secretory proteins, disulfide bonds cross-linking specific cysteines are added to stabilize a protein or to join covalently different polypeptides. These bonds are crucial for the stability of the final protein structure, thus mispairing of cysteine residues can prevent proteins from attaining their native conformation and lead to misfolding [15].
Typical PDI is the most prominent member of a family of related proteins (PDI-like) characterised by one, two or three thioredoxin-like active domains [16]. It has been cloned and sequenced in many plant species, such as alfalfa [17], barley [18], maize [19], castor bean [20], soyabean [21-24], common and durum wheat [25,26]. A detailed knowledge of the complexity and diversity of the genes encoding PDI and PDI-like proteins in Arabidopsis thaliana, wheat and other plant species was described by [27,28].
Wheat genes coding for typical PDI in bread wheat have been located in homoeologous chromosome group four [29]. Analyses performed on 23 species of Triticum and Aegilops [30], indicated that PDI restriction fragments were highly conserved within each species and confirmed that plant PDI is encoded either by one or few copy sequences, respectively in diploid and polyploid species. The nucleotide sequences of the three genes located respectively on genomes A, B, and D (designed as GPDI-4A, GPDI-4B and GPDI-4D) were 3561 bp, 3527 bp and 3466 bp long. The comparison of typical PDI gene sequences of wheat, rice and Arabidopsis showed a significant conservation of the exon/intron structure across the three species [31]. More detailed study on the complexity and diversity of the genes encoding PDI and PDI-like proteins in wheat and other plant species also showed a significant conservation of the exon/intron structure [28].
The expression analysis of the typical PDI homoeologous genes located on chromosomes 4A, 4B and 4D of bread wheat cv. Chinese Spring (CS) (Ciaffi et al., 2006) showed that the PDI transcripts, although constitutively present at a low level in all the analyzed tissues, are equally abundant in the developing caryopses but are differentially expressed in spikelets, roots and leaves [25, 31]. The PDI-4A transcription was higher in spikelets, that of PDI-4B were higher in roots while the PDI-4 D transcripts were more abundant in leaves. The transcripttion levels of the three genes were higher in the early stage of seed development (6 - 14 DAA) and decreased during middle to late stage of (18 - 34 DAA) grain filling [31]. Within the upstream putative promoter region of the three homoeologous genes cloned from bread wheat cv CS, respectively 1352 bp for PromPDI-4A, 1370 bp for PromPDI-4B and 1292 bp for PromPDI-4D long, several cis-acting elements involved in endosperm specific expression were detected, consistently with the higher PDI expression detected in the kernels. PDI promoter sequence analysis of Triticum urartu, Aegilops speltoides and Aegilops tauschii was given by [32].This paper reports variability in a ~700 bp region, comprising ~600 bp of the 5’ upstream putative promoter region and 88 bp of the first exon of the typical PDI gene from the diploid species Triticum urartu (AA) Aegilops speltoides (BB) and Aegilops tauschii (DD). For simplicity we will further refer to the amplified ~700 bp region shortly as the ~700 bp target region.
2. Materials and Methods
2.1. Plant Material and DNA Extraction
A total of 15 accessions, 5 each from Triticum urartu (AA), Aegilops speltoides (BB) and Aegilops tauschii (DD) were used in this study (Table 1). Eight plants per accessions were grown and analysed for a total of 120 plants. Flag leaves were collected at heading stage from plants grown in green house (January-June 2008), immediately frozen in liquid nitrogen and kept at −80˚C until use. About 200 mg of leaf tissue was ground in liquid nitrogen and genomic DNA was extracted using Sigma Gen Elute Plant Genomic DNA Kit (G2N-350, Sigma Aldrich, St. Louis, Mo.).
2.2. Primers Design and PCR Amplification
Primers were designed on the basis of the known homoeologous PDI promoter and gene sequences isolated from bread wheat cv. Chinese Spring (Ciaffi et al., 2006), using DNAMAN 4.15 program. Putative promoters of typical PDI were amplified by using the following pri mer pairs PDIAPDF2-PDIAPDR1 and PDIAPDF5- PDIAPDR6 for A Genome, PDIAPDF2-PDIAPDR1 and PDIAPDF2-PDIAPDR5 for B genome and PDIAPDF2- PDIAPDR1 and PDIAPDF3-PDIAPDR5 for D genome, cloned and sequenced in one accession for each wild

Table 1. Accession numbers and geographical origin of the wheat samples used in this study.
species. Further amplification reactions were carried out in the 120 plants using the primer pair PDIAPDF2- PDIAPDR1 and directly sequenced using the PDIAPDF2 primer (Table 2).
PCR reaction mixture included 10 ng/μl genomic DNA, 0.20 mM dNTPs 5 ul, 0.05 units/µl of Taq 0.50 µl (go-taq, Promega), 0.40 μM primer 2.0 µl (0.20 µM per each), 5× buffer 10 µl and 25.5 µl of ddH2O used per reaction (final volume of 50 μl). PCR condition included initial denaturation step at 95˚C for 3 min, then 32 cycles of (95˚C for 1min, 59˚C for 35 sec, 72˚C for 1 min) and final elongation at 72˚C for amplification of the sequences from the A, B and D Genome.
2.3. Cloning and Sequencing
One conserved primer pair (PDIAPDF2-PDIAPDR1) and one genome specific primer pair (Table 2) were used to amplify, clone and sequence the DNA extracted from one plant each of the 3 wild relatives. PCR products of expected size were excised from the gel, purified using the High Pure Purification kit (Roche) according to manufacturer’s instructions, and cloned into the pGEM-T easy plasmid vector (Promega). Plasmids were transformed by heat shock into Escherichia coli strain DH5α. Bacteria were plated onto LB medium containing ampicillin, XGal and IPTG, and recombinant plasmids were identified by blue/white screening. For each primer combination two independent PCR reactions were performed and a total of 6 clones were sequenced. Plasmid DNA for sequencing reaction was prepared from 3 ml overnight cultures using a plasmid miniprep kit (Qiagen). Sequencing was performed on both strands by the ABI PRISM 377 capillary sequencer (PE Applied Biosystem) using an ABI Prism Dye Terminator sequencing kit (PE Applied Biosystem) and sequenced both with vector and sequence specific primers. The target region was further analysed in all 120 plants using the conserved primer pair PDIAPDF2-PDIAPDR1 and directly sequenced.
2.4. Data Analysis
Sequences obtained from Sequencer were analysed in Chromas version 2.3 software (http://technelysium.com. au/chromas.html) to identify any unresolved bases and subjected to final visual inspection. Analysis of 120

Table 2. List of primers used for the isolation of PDI partial promoter analysis.
sequences revealed that plants within accessions of all three wild species were identical. So among 120 sequences analyzed, a total of 15 sequences (one for each accession) from the three wild species (5 accessions each × 3 wild species = 15 sequences) were further used analyses along with the 3 homoeologous sequences from CS obtained from NCBI database (GPDI-4A, AJ868102; GPDIA-4B, AJ868103; GPDI-4D, AJ868104; [31]). So totally 18 sequences were used for multiple alignment and other analysis. The 18 sequences were multiple aligned with Clustal X software version 2.011 [33], IUB as DNA weight matrix with default parameters. A phylogenetic tree was constructed on obtained data by using the neighbour-joining (NJ) method [34], using MEGA version 4 with neighbour joining option and 1000 bootstrap [35]. Two measures of nucleotide polymorphism were calculated [36]: nucleotide diversity (π) based on the average number of pairwise comparisons in a sample [37] and Watterson’s estimator (θ) based on the number of segregating sites [38] using DnaSP v5 soft-ware [39]. Sequences obtained from this work were submitted in EMBL Nucleotide Sequence Database with sequence ID starting from FN677020 to FN677037.
3. Results
3.1. Phylogenetic Analysis
The evolutionary relationship between 18 sequences, comprising part of the putative promoter as well as part of the first exons was studied by phylogeny reconstructtion. These included 5 sequences each from Triticum urartu, Aegilops speltoides, Aegilops tauschii for a total of 15 new sequences and the three homoeologous sequences from CS previously reported by [31]. The phylogenetic tree was constructed using three different methods namely the neighbour-joining (NJ) method, Minimum Evolution (ME) and Maximum Parsimony (MP) methods. As the results of the three methods were similar, only NJ tree is presented here (Figure 1).
All sequences were grouped into two major clusters, one containing the sequences derived from the A genome, and one containing two subcluster each formed respectively by the B and D genome sequences. The sequences from Aegilops speltoides (BB) closely clustered together with the sequence located on the chromosome 4B of CS. Aegilops speltoides is native to the eastern Mediterranean and Middle East region and consists of two sub species, sub species speltoides (awnless lemma except for apical spikelet) and sub species ligustica (awned lemma) [40, 41]. Both sub species were present in this study. Sequences from accessions IG48766 (Aegilops speltoides var speltoides, Lebanon) IG46812 (Aegilops speltoides var ligustica, Turkey), IG46811 (Aegilops speltoides var speltoides, Turkey), IG46593 (Aegilops speltoides var ligustica, Syria) and IG46597 (Aegilops speltoides var speltoides, Syria) were all similar but not identical and clustered independently. The homoeologous B genome of hexaploid wheat is closely related to the genome of Aegilops speltoides that is proposed to be its wild progenitor [1,11,42].
The sequences from the Aegilops tauschii (DD) accessions closely clustered with that located on the 4D chromosome of CS and in particular three accessions from Iran, AE525 (Aegilops tauschii Coss. subsp. strangulate), AE526 (Aegilops tauschii Coss) and AE527 (Aegilops tauschii Coss), presented 100% sequence identity in the target region and clustered therefore together. 100% sequence identity characterized also the remaining two accessions AE1068 (Syria) and AE541 (Iran) which formed a second subcluster. Several studies underline that Aegilops tauschii, also known as wild goat grass from the Middle East, crossed with modern ver-

Figure 1. Phylogenetic tree of PDI gene sequences (partial promoter and partial part of first exon) of Triticum urartu (AA), Aegilops speltoides (BB) and Aegilpos taushcii (DD).
sions of emmer wheat to produce bread wheat [40,43, 44].
As expected also all the accessions of Triticum urartu (AA) closely clustered with the sequence from CS located on chromosome 4A. The accessions from Lebanon (IG45475), Turkey (IG45108), Jordan (IG115817) and Syria (IG44831) formed a single group with 100% sequence identity, the accession from Iran (IG45477) stayed separately.
3.2. Test for Neutrality
For test of neutrality, level of nucleotide diversity and population mutation, as estimated by π [37] and θ [38], were evaluated for deviation from an equilibrium neutral model. To test for the operation of natural selection in the region under study, we applied following neutrality tests [45,46] potentially able to detect deviations of the observed data from the expectations under the neutral model. Neutrality test conducted on the 5 sequences from each Triticum urartu, Aegilops speltoides and Aegilops tauschii showed high level of nucleotide diversity and Watterson estimator value (π = 0.00954, θ = 0.01110) for Aegilops speltoides when compared to Triticum urartu (π = 0.00112, θ = 0.00135) and Aegilops taushcii (π = 0.00168, θ = 0.00135) (Table 3). Neutrality tests such as Tajima’s D Fu, Li’s D and Fu and Li’s D gave negative values for accessions from Triticum urartu and Aegilops speltoides and positive values for Aegilops taushcii but none of the values resulted significant (Table 3). Haplotype diversity observed in Triticum urartu, Aegilops speltoides and Aegilops tauschii sequences was 0.400, 1.000 and 0.600 respectively.
4. Discussion
It is well known that cultivated wheat has formed from throughout several polyploidization processes [43]. Phylogenetic analysis of 18 sequences showed a genome specific clustering of the analysed sequences.
Studies of haplotype diversity in plants are limited and so far restricted to a few species, such as Arabidopsis, soybean, barley, and maize [47-51]. Haplotype diversity observed in Triticum urartu, and Aegilops tauschii sequences was low (0.400, and 0.600), when compared with that of Aegilops speltoides (1.000). Intraspecific variation at the haplotype level has been studied to varying degrees in different plant species. The most comprehensive data are available for Arabidopsis (Arabidopsis thaliana), in which oligonucleotide resequencing arrays allowed comparison of multiple ecotypes at the whole-genome level and showed that nucleotide substitutions are irregularly distributed across the genome and that about 4% of genomic sequences were absent in some ecotypes relative to the reference genome [52,53]. Additionally, regions with almost no sequence
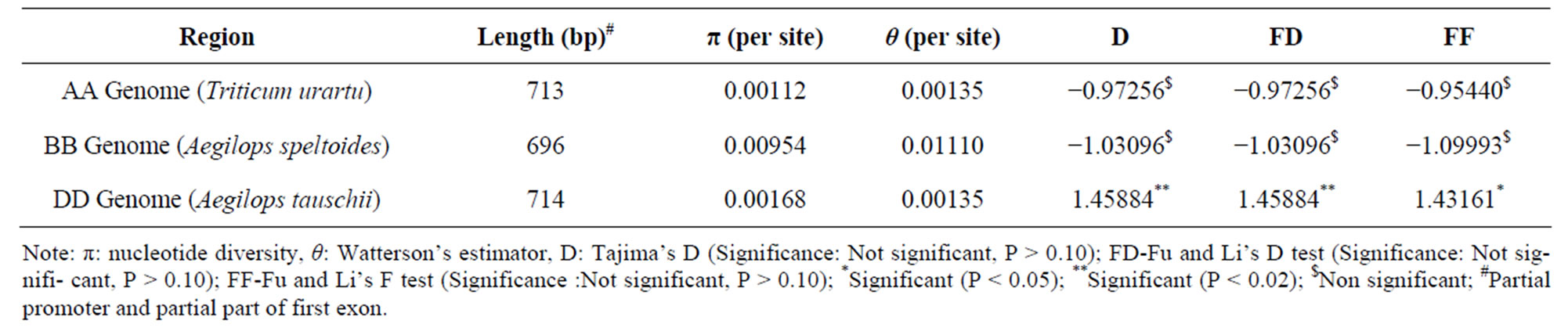
Table 3. Estimates of nucleotide diversity and test of statistics for selection in AA Genome (Triticum urartu), BB Genome (Aegilops speltiodes) and DD Genome (Aegilops tauschii).
diversity were interpreted as results of recent selective sweeps [52].
Neutrality tests [Tajima’s D, [46]; Fu and Li’s D, [45]; Fu and Li F, [45] conducted on Triticum urartu, Aegilops speltoides and Aegilops tauschii showed negative values for accessions from Triticum urartu and Aegilops speltoides and positive values for Aegilops taushcii accessions, but none of them were significant. Similar results were obtained in Mustard Genus (Leavenworthia) for the (cytosolic phosphoglucose isomerase) PgiC Locus [54].
5. Conclusion
Former studies in wheat had been restricted to the characterization of the genes encoding the typical PDI and their promoter, and the cloning and characterization of the complete set of genes encoding PDI and PDI like proteins in bread wheat (Triticum aestivum cv. Chinese Spring) and the comparison of their sequence, structure and expression with homologous genes from other plant species, which is of special interest for its involvement in determining the bread making qualities and technological properties of flour. The interest of extending the study to promoters of wild species is due to high conservation of gene, due to their relevant metabolic functions, as well as to the interesting expression pattern found in previous work. This study did not detect any variation within accession in all the three genomes studied. This might be due to the genetic variation in gene bank accessions does not reflect the genetic diversity of their original population. It would be more useful if further studies involve large number of accessions and less number of plants per accessions. Future studies should involve characterisation of proximal and distal end of the Typical PDI promoter in diploid and tetraploid progenitor from diverse geographic origin of world to determine the diversity among larger number of accessions.
6. Acknowledgements
I thank Professor Enrico Porceddu and Dr. Elisa Laura di Aloisio for reviewing this manuscript. I thank Shalini Narasimhan for helping me in editing and finalizing the manuscript.
REFERENCES
- B. S. Gill, R. Appels, A. M. Botha-Oberholster, C. R. Buell, J. L. Bennetzen, B. Chalhoub, F. Chumley, J. Dvorák, M. Iwanaga, B. Keller, W. Li, W. R. McCombie, Y. Ogihara, F. Quetier and T. Sasaki, “A Workshop Report on Wheat Genome Sequencing: International Genome Research on Wheat Consortium,” Genetics, Vol. 168, No. 2, 2004, pp. 1087-1096. doi:10.1534/genetics.104.034769
- H. Kihara, “Uber Cytolosgische Studien bei Einigen Getreidearten. I. Spezies-Bastarde des Weizens und Weizenmggen-Bastarde,” Botanical Magazine (Tokyo), Vol. 33, 1919, pp. 17-38.
- H. Kihara, “Cytologische und Genetische Studien bei Wichtigen Getreidearten mit Besonderer Rucksicht auf das Verhalten der Chromosomen und die Sterilität in den Bastarden,” Memoirs of the College of Science, Kyoto Imperial University, Vol. 8, 1924, pp. 1-200.
- K. Sax, “Sterility in Wheat Hybrids. II. Chromosome Behavior in Partially Sterile Hybrids,” Genetics, Vol. 7, No. 6, 1922, pp. 49-68.
- R. Morris and E. R. Sears, “The Cytogenetics of Wheat and Its Relatives,” In: K. S. Quisenberry and L. P. Reitz, Eds., Wheat and Wheat Improvement, American Society of Agronomy, Madison, 1967, pp. 19-87.
- B. R. Baum and L. G. Bailey, “The Origin of the A Genome Donor of Wheats (Triticum: Poaceae): A Perspective Based on the Sequence Variation of the 5S DNA Gene Units,” Genetic Resources and Crop Evolution, Vol. 51, No. 2, 2004, pp. 183-196. doi:10.1023/B:GRES.0000020861.96294.f4
- J. Dvorak, P. Tetizi, H. B. Zhang and P. Resta, “The Evolution of Polyploid Wheats: Identification of the A Genome Donor Species,” Genome, Vol. 36, No. 1, 1993, pp. 21-31.doi:10.1139/g93-004
- M. Feldman, “Origin of Cultivated Wheat,” In: A. P. Bonjean and W. J. Angus, Eds., The World Wheat Book: A History of Wheat Breeding, Intercept Ltd., London, 2001, pp. 3-56.
- J. Jiang and B. S. Gill, “Different Species-Specific Chromosome Translocations in Triticum timopheevi and Triticum turgidum Support the Diphyletic Origin of Polyploid Wheats,” Chromosome Research, Vol. 2, No. 1, 1994, pp. 59-64. doi:10.1007/BF01539455
- H. Kihara, “Discovery of the DD-Analyser, One of the Ancestors of Triticum vulgare,” Agricuture and Horticulture, Vol. 19, 1944, pp. 889-890.
- J. Dvorak, M. C. Luo and Z. L. Yang, “Genetic Evidence on the Origin of T. aestivum L.,” In: A. Damania, Ed., The Origins of Agriculture and the Domestication of Crop Plants in the Near East, ICARDA, Aleppo, 1998, pp. 235-251.
- S. Chao, W. Zhang, E. Akhunov, J. Sherman, Y. Ma, M. C. Luo and J. Dubcovsky, “Analysis of Gene-Derived SNP Marker Polymorphism in US Wheat (Triticum aestivum L.) Cultivars,” Molecular Breeding, Vol. 23, No. 1, 2009, pp. 23-33. doi:10.1007/s11032-008-9210-6
- P. R. Shewry, J. A. Napier and A. S. Tatham, “Seed Storage Proteins: Structure and Biosynthesis,” Plant Cell, Vol. 7, No. 7, 1995, pp. 945-956.
- M. Schwaller, B. Wilkinson and H. F. Gilbert, “Reduction-Reoxidation Cycles Contribute to Catalysis of Disulfide Isomerisation by Protein-Disulfide Isomerase,” Journal of Biological Chemistry, Vol. 278, No. 9, 2003, pp. 7154-7159.doi:10.1074/jbc.M211036200
- B. P. Tu and J. S. Weissman, “Oxidative Protein Folding in Eukaryotes: Mechanisms and Consequences,” Journal of Cell Biology, Vol. 164, No. 3, 2004, pp. 341-346. doi:10.1083/jcb.200311055
- D. M. Ferrari and H. D. Soling, “The Protein DisulphideIsomerase Family: Unravelling a String of Folds,” Biochemistry Journal, Vol. 339, No. 1, 1999, pp. 1-10. doi:10.1042/0264-6021:3390001
- B. S. Shorrosh and R. A. Dixon, “Molecular Cloning of a Putative Plant Endomembrane Protein Resembling Vertebrate Protein Disulfide-Isomerase and a Phosphatidylinositol-Specific Phospholinase,” Proceedings of National Academy of the Sciences, Vol. 88, No. 23, 1991, pp. 10941-10945.
- F. Chen and P. M. Hayes, “Nucleotide Sequence and Developmental Expression of Duplicated Genes Encoding Protein Disulfide Isomerase in Barley (Hordeum vulgare L.),” Plant Physiology, Vol. 106, No. 4, 1994, pp. 1705-1706. doi:10.1104/pp.106.4.1705
- C. P. Li and B. A. Larkins, “Expression of Protein Disulfide Isomerase Is Elevated in the Endosperm of the Maize Floury-2 Mutant,” Plant Molecular Biology, Vol. 30, No. 5, 1996, pp. 873-882. doi:10.1007/BF00020800
- S. J. Coughlan, C. Hastings and R. J. Winfrey, “Molecular Characterization of Plant Endoplasmic Reticulum: Identification of Protein Disulfide-Isomerase as the Major Reticuloplasmin,” European Journal of Biochemistry, Vol. 235, No. 1-2, 1996, pp. 215-224. doi:10.1111/j.1432-1033.1996.00215.x
- K. Iwasaki, S. Kamauchi, H. Wadahama, M. Ishimoto, T. Kawada and R. Urade, “Molecular Cloning and Characterization of Soybean Protein Disulfide Isomerase Family Proteins with Non Classic Active Center Motifs,” FEBS Journal, Vol. 276, No. 15, 2009, pp. 4130-4141. doi:10.1111/j.1742-4658.2009.07123.x
- S. Kamauchi, H. Wadahama, K. Iwasaki, Y. Nakamoto, K. Nishizawa, M. Ishimoto, T. Kawada and R. Urade, “Molecular Cloning and Characterization of Two Soybean Protein Disulfide Isomerases as Molecular Chaperones for Seed Storage Proteins,” FEBS Journal, Vol. 275, No. 10, 2008, pp. 2644-2658. doi:10.1111/j.1742-4658.2008.06412.x
- H. Wadahama, S. Kamauchi, M. Ishimoto, T. Kawada and R. Urade, “Protein Disulfide Isomerase Family Proteins Involved in Soybean Protein Biogenesis,” FEBS Journal, Vol. 274, No. 3, 2007, pp. 687-703. doi:10.1111/j.1742-4658.2006.05613.x
- H. Wadahama, S. Kamauchi, Y. Nakamoto, K. Nishizawa, M. Ishimoto, T. Kawada and R. Urade, “A Novel Plant Protein Disulfide Isomerase Family Homologous to Animal P5: Molecular Cloning and Characterization as a Functional Protein for Folding of Soybean Seed Storage Proteins,” FEBS Journal, Vol. 275, No. 3, 2008, pp. 399-410. doi:10.1111/j.1742-4658.2007.06199.x
- M. Ciaffi, A. R. Paolacci, L. Dominici, O. A. Tanzarella and E. Porceddu, “Molecular Characterization of Gene Sequences Coding for Protein Disulfide Isomerase (PDI) in Durum Wheat (Triticum turgidum ssp. durum),” Gene, Vol. 265, No. 1-2, 2001, pp. 147-156. doi:10.1016/S0378-1119(01)00348-1
- Y. Shimoni, G. Segal, X. Zhu and G. Galili, “Nucleotide Sequence of a Wheat cDNA Encoding Protein Disulfide Isomerase,” Plant Physiology, Vol. 107, No. 1, 1995, pp. 281. doi:10.1104/pp.107.1.281
- N. L. Houston, C. Fan, Q. Y. Xiang, J. M. Schulze, R. Jung and R. S. Boston, “Phylogenetic Analyses Identify 10 Classes of the Protein Disulfide Isomerase Family in Plants, Including Single-Domain Protein Disulfide Isomerase-Related Proteins,” Plant Physiology, Vol. 137, No. 2, 2005, pp. 762-778. doi:10.1104/pp.104.056507
- E. d'Aloisio, A. R. Paolacci, A. P. Dhanapal, O. A. Tanzarella, E. Porceddu and M. Ciaffi, “The Protein Disulfide Isomerase Gene Family in Bread Wheat (T. aestivum L.),” BMC Plant Biology, Vol. 10, 2010, pp. 101. doi:10.1186/1471-2229-10-101
- M. Ciaffi, L. Dominici, O. A. Tanzarella and E. Porceddu, “Chromosomal Assignment of Gene Sequences Coding for protein Disulphide Isomerase (PDI) in Wheat,” Theoritical and Applied Genetics, Vol. 98, No. 3-4, 1999, pp. 405-410. doi:10.1007/s001220051086
- M. Ciaffi, L. Dominici, E. Umana, O. A. Tanzarella and E. Porceddu, “Restriction Fragment Length Polymorphism (RFLP) for Protein Disulfide Isomerase (PDI) Gene Sequences in Triticum and Aegilops Species,” Theoritical and Applied Genetics, Vol. 98, No. 1-2, 2000, pp. 220- 226. doi:10.1007/s001220051472
- M. Ciaffi, A. R. Paolacci, E. d’Aloisio, O. A. Tanzarella and E. Porceddu, “Cloning and Characterization of Wheat PDI (Protein Disulfide Isomerase) Homoeologous Genes and Promoter Sequences,” Gene, Vol. 366, No. 2, 2006, pp. 209-218. doi:10.1016/j.gene.2005.07.032
- A. P. Dhanapal, M. Ciaffi, E. Porceddu and E. d’Aloisio, “Protein Disulphide Isomerase Promoter Sequence Analysis of Triticum urartu, Aegilops speltoides and Aegilops tauschii,” Plant Genetic Resources, Vol. 9, No. 2, 2011, pp. 338-341. doi:10.1017/S147926211100013X
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, “Clustal W and Clustal X Version 2.0,” Bioinformatics, Vol. 23, No. 21, 2007, pp. 2947-2948. doi:10.1093/bioinformatics/btm404
- N. Saitou and M. Nei, “The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees,” Molecular Biology and Evolution, Vol. 4, No. 4, 1987, pp. 406-425.
- K. Tamura, J. Dudley, M. Nei and S. Kumar, “MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0,” Molecular Biology and Evolution, Vol. 24, No. 8, 2007, pp. 1596-1599. doi:10.1093/molbev/msm092
- F. Tajima, “Evolutionary Relationship of DNA Sequences in Finite Populations,” Genetics, Vol. 105, No. 2, 1983, pp. 437-460.
- M. Nei, “Molecular Evolutionary Genetics,” Columbia University Press, New York, 1987.
- G. A. Watterson, “On the Number of Segregating Sites in Genetic Models without Recombination,” Theoretical Population Biology, Vol. 7, No. 2, 1975, pp. 256-276. doi:10.1016/0040-5809(75)90020-9
- P. Librado and J. Rozas, “DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data,” Bioinformatics, Vol. 25, No. 11, 2009, pp. 1451-1452. doi:10.1093/bioinformatics/btp187
- G. Kimber and M. Feldman, “Wild Wheat, an Introduction,” College of Agriculture University of Missouri Special Report, Vol. 353, Columbia, 1987, p. 146.
- M. van Slageren, “Wild Wheats: A Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae),” Wageningen Agriculture University Papers, Vol. 7, 1994, pp. 513.
- J. Dvorak, M. C. Luo, Z. L. Yang and H. B. Zhang, “The Structure of the Aegilops tauschii genepool and the Evolution of Hexaploid Wheat,” Theoritical and Applied Genetics, Vol. 97, No. 4, 1988b, pp. 657-670.
- P. Gitte, O. Seberg, M. Yde and K. Berthelsen, “Phylogenetic Relationships of Triticum and Aegilops and Evidence for the Origin of the A, B, and D Genomes of Common Wheat (Triticum aestivum),” Molecular Phylogenetics and Evolution, Vol. 39, No. 1, 2006, pp. 70-82. doi:10.1016/j.ympev.2006.01.023
- T. Simonite, “Ancient Genetic Tricks Shape Up Wheat Turning Back the Evolutionary Clock Offers Better Crops for Dry Regions,” Nature, 2006.
- Y. X. Fu and W. H. Li, “Statistical Tests of Neutrality of Mutations,” Genetics, Vol. 133, No. 3, 1993, pp. 693-709.
- E. Tajima, “Statistical Test for Testing the Neutral Mutation Hypothesis by DNA Polymorphism,” Genetics, Vol. 123, No. 3, 1989, pp. 585-595.
- D. Charlesworth, C. Bartolome, M. H. Schierup and B. K. Mable, “Haplotype Structure of the Stigmatic Self-Incompatibility Gene in Natural Populations of Arabidopsis Iyrata,” Molecular Biology and Evolution, Vol. 20, No. 11, 2003, pp. 1741-1753. doi:10.1093/molbev/msg170
- N. C. Collins, T. Lahaye, C. Peterhansel, A. Freialdenhoven, M. Corbitt and P. Schulze-Lefert, “Sequence Haplotypes Revealed by Sequence-Tagged Site Fine Mapping of the Ror1 Gene in the Centromeric Region of Barley Chromosome 1H,” Plant Physiology, Vol. 125, No. 3, 2001, pp. 1236-1247. doi:10.1104/pp.125.3.1236
- M. Nordborg, J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory, J. Hagenblad, M. Kreitman, J. N. Maloof, T. Noyes, P. J. Oefner, E. A. Stahl and D. Weigel, “The Extent of Linkage Disequilibrium in Arabidopsis thaliana,” Nature Genetics, Vol. 30, No. 2, 2002, pp. 190-193. doi:10.1038/ng813
- A. Rafalski and M. Morgante, “Corn and Humans: Recombination and Linkage Disequilibrium in Two Genomes of Similar Size,” Trends in Genetics, Vol. 20, No. 2, 2004, pp. 103-111. doi:10.1016/j.tig.2003.12.002
- Y. L. Zhu, Q. J. Song, D. L. Hyten, C. P. Van Tassell, L. K. Matukumalli, D. R. Grimm, S. M. Hyatt, E. Fickus, N. D. Young and P. B. Cregan, “Single-Nucleotide Polymorphisms in Soybean,” Genetics, Vol. 163, No. 3, 2003, pp. 1123-1134.
- R. M. Clark, G. Schweikert, C. Toomajian, S. Ossowski, G. Zeller, P. Shinn, N. Warthmann, T. Hu, G. Fu, D. A. Hinds, H. Chen, K. A. Frazer, D. H. Huson, B. Schölkopf, M. Nordborg, G. Rätsch, J. R. Ecker and D. Weigel, “Common Sequence Polymorphisms Shaping Genetic Diversity in Arabidopsis thaliana,” Science, Vol. 317, No. 5836, 2007, pp. 338-342. doi:10.1126/science.1138632
- G. Zeller, R. M. Clark, K. Schneeberger, A. Bohlen, D. Weigel and G. Rätsch, “Detecting Polymorphic Regions in Arabidopsis thaliana with Resequencing Microarrays,” Genome Research, Vol. 18, No. 6, 2008, pp. 918-929. doi:10.1101/gr.070169.107
- D. A. Filatov and D. Charlesworth, “DNA Polymorphism, Haplotype Structure and Balancing Selection in the Leavenworthia PgiC Locus,” Genetics, Vol. 153, No. 3, 1999, pp. 1423-1434.

