Advances in Microbiology
Vol.04 No.13(2014), Article ID:50652,10 pages
10.4236/aim.2014.413104
Nitrogen Source Affects Glycolipid Production and Lipid Accumulation in the Phytopathogen Fungus Ustilago maydis
Ariana Zavala-Moreno1, Roberto Arreguin-Espinosa2, Juan Pablo Pardo3, Lucero Romero-Aguilar1, Guadalupe Guerra-Sánchez1*
1Departamento de Microbiología, Escuela Nacional de Ciencias Biológicas, IPN, México D.F., Mexico
2Departamento de Química de Biomacromoléculas, Instituto de Química, UNAM, Ciudad Universitaria, México D.F., Mexico
3Departamento de Bioquímica, Facultad de Medicina, UNAM, Ciudad Universitaria, México D.F., Mexico
Email: *lupegs@hotmail.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 3 August 2014; revised 2 September 2014; accepted 5 October 2014
ABSTRACT
When cultured in medium limited of nitrogen sources, the phytopathogen Ustilago maydis produces two amphipathic glycolipids: Ustilagic acid (UA) and Mannosylerythritol lipid (MEL), which in addition to the hydrophilic moiety, contain di- or tri-hydroxylated C16 fatty acids (UA), or C8 and C16 saturated fatty acids (MEL). We compared the growth and morphology of cells in YPD and in minimum media containing glucose and nitrogen sources such as nitrate or urea and those deprived of nitrogen. Nitrogen-starved cells showed a dramatic accumulation of internal lipids identified as lipid droplets when stained with the hydrophobic probe BODIPY; these lipid droplets were enriched in unsaturated fatty acids. Fatty acids in YPD or medium containing nitrate as nitrogen source showed a combination of saturated/unsaturated lipids, but when urea was the nitrogen source, cells only contained saturated fatty acids. The glycolipid profiles produced in the presence or absence of nitrogen showed preferences towards the production of one kind of glycolipid: cells in media containing nitrate or urea produced different proportions of UA/MEL, but under nitrogen starvation cells contained only UA. The emulsification capacity of the glycolipids pro- duced in media with or without nitrogen was similar (72% - 76%). HPLC of the glycolipids allowed the separation of fractions with different emulsifying characteristics. Our results indicate that U. maydis accumulates lipid droplets when deprived of nitrogen source and confirm that UA is not under nitrogen control, but rather that MEL and lipid droplets are produced and oppositely regulated by nitrogen.
Keywords:
Nitrogen Starvation, Yeast Glycolipids, Lipid Droplets, Lipid Accumulation

1. Introduction
The yeast of Ustilago maydis produces extracellular glycolipids with surfactant activity that enhance the availability of hydrophobic nutrients during its interaction with the host [1] [2] . These biosurfactants are derivatives of two classes of amphiphatic glycolipids: Ustilagic acid (UA) and Mannosylerythritol lipids (MEL). They are secreted into the extracellular media at the stationary phase when nitrogen becomes an environmental limitation [3] . Their surfactant properties make this type of glycolipids very promising for commercial production in cosmetic and pharmaceutical industry [4] - [6] . In recent years, several reports described enhanced yields of these biosurfactants when U. maydis cells are grown in yeast hydrolysates, urea and nitrate or ammonium salts, as nitrogen sources, and using also multiple carbon sources such as glucose, sucrose, lipids and glycerol [7] - [9] . Different strains have been used for the production of glycolipids and the yield of the two glycolipids may be affected by the availability of nitrogen source [10] . The biosynthesis of UA and MEL depends on specific gene clusters [1] [11] [12] . For UA, cyp1 and cyp2 genes are required for terminal and subterminal hydroxylation of hexadecanoic acid [12] . Mannosylerythritol lipids (MEL) are secreted in at least four different varieties (ABCD), depending on the number of acetyl groups attached to the disaccharide [3] - [11] and a glycosyltransferase is essential for the biosynthesis of MEL; expression of this enzyme is strongly induced by limitations on nitrogen availability [11] . To produce these glycolipids, U. maydis requires raw materials to synthesize the lipid and carbohydrate moieties. Different carbon sources have been used in an effort to attain high yields of these glycolipids [3] - [9] , but less attention has been placed on identifying the effect of nitrogen sources on the production of glycolipids. When cells are limited in terms of their nitrogen sources, neutral lipids are accumulated as discrete deposits, named lipid droplets (LD) [13] [14] . Moreover, when cells are switched from a rich media to a minimal medium lacking of nitrogen source, autophagy is induced to cope with the starvation condition [15] . In this work, we studied the growth and morphology of U. maydis yeasts cultured in YPD and minimal media (MM) containing glucose as carbon source without a nitrogen source (MM−N), or glucose under nitrogen limiting conditions using nitrate (MM+N) or Urea (MM+U) as nitrogen sources. We also reported the presence of lipid droplets, accumulation of unsaturated fatty acids and production of the biosurfactant Ustilagic acid (UA) in cells deprived of nitrogen. Glycolipids profile was altered by the presence of nitrogen in the culture media, with the production of mixtures of UA and MEL, and changes in the saturated/unsaturated fatty acids ratio. The emulsifying capacity of glycolipids synthesized by cells grown either under nitrogen limiting conditions (nitrate or urea) or deprived of nitrogen was also evaluated. This study confirmed that in contrast to MEL, UA is not under nitrogen control, and that U. maydis is a potential microorganism for the production of lipids for biofuels and biosurfactants in nitrogen deprived conditions.
2. Materials and Methods
2.1. Chemicals
All reagents were reactive or HPLC grade from JT Baker or Sigma Chem Co. Surfactin from Bacillus subtilis was purchased from Sigma. BODIPY (4, 4-difluoro-3a, 4a,-diaza-s-indacene) 483/503 was from Invitrogen and prepared as a 10 mM stock solution in DMSO and kept at −70˚C. For assay of total lipids, a clinical kit was obtained from Spinreact. TLC plates of Silica-Fluorescent gel 60 were from Merck.
2.2. Strain and Culture Condition
The Ustilago maydis ATCC 201384 FB2 strain was obtained from American Type Culture Collection and kept in 25% glycerol at −70˚C. Cells were streaked in YPD plates (10 g∙L−1 glucose, 1.5 g∙L−1 ammonium nitrate, 2.5 g∙L−1 peptone, 10 g∙L−1 yeast extract and 62.5 ml∙L−1 of salt solution adjusted to pH 7.0 and agarose 2%), and incubated at 28˚C for 24 h. They were subsequently transferred to 100 ml YPD media and shaken at 125 rpm at 29˚C by 24 h. This pre-culture was used as inoculum for all further experiments. Minimum media with nitrate (MM+N) contained 0.3% potassium nitrate, 1% glucose and 62.5 ml∙L−1 of salts solution [16] , adjusted to pH 7.0 with 1 N NaOH. The minimum media without a nitrogen source (MM−N) contained 1% glucose and salts solution at pH 7.0. Minimum media with urea (MM+U) contained 1% glucose, 0.06% urea and same salts solution. The growth curve was determined by inoculating 50 OD600 in 1 L and incubated for 7 days in a rotary shaker (125 rpm) at 29˚C. Aliquots of 1 ml were taken to measure the growth of cells by optical density at 600 nm and the morphology of cells was monitored by light microscopy (100×) in 6 - 12 h time intervals during the culture period. Statistical analysis was performed using Excel 2007. Data was analyzed using overall one-way analysis of variance (ANOVA) and the differences observed were significant at P ≤ 0.05.
2.3. Fluorescence Microscopy of Lipid Droplets in U. maydis Cells
For the determination of lipid droplets by fluorescense microscopy, aliquots were withdrawn during growth and immediately treated as described in [17] . Briefly, aliquots of 5 ml of cellular suspension (10 OD600) were fixed with 24% formaldehyde, centrifugated at 2500 g and washed with H2O milliQ (2×). Cells were re-suspended at 10U OD and 10 µl were mixed with 200 µl of 50 mM KI and 5 µl of BODIPY. Cells were mounted onto a polylysine treated glass slide. Microscopy was performed on Olympus 1× 81 at 100× oil immersion objective. The captured images were analyzed using Image software.
2.4. Measurement of Total Lipids and Identification by GC/MS
Cultures were grown in the media of YPD, MM+N, MM−N and MM+U and cells harvested by centrifugation at 2500 g for 10 min. 40 mg of cells from each media (wet weight) were re-suspended in 300 µl of 50 mM HEPES buffer pH 7.0 and equal mg of glass beads (0.5 mm, Biospec) were added and lyzed by vortexing for 10 cycles of 1 min at 4˚C. Lysed cells were separated by centrifugation at 2500 g for 15 min and the glass beads were washed with 100 μl of the same HEPES buffer and spun. The total lysate was centrifuged at the same condition to get the supernatant. Lipids were extracted using the final supernatant and mixed with chloroform/methanol/ water (65:25:4) solution. After shaking for 5 minutes, the organic phase was separated by centrifugation and the solvent was evaporated under vacuum. Fatty acids were weighed and re-suspended in 1 ml of methanol. Aliquots of 5 µl were taken to analyze by gas chromatograph with a quadruple mass spectrometer in a JEOL GC mate with an EI+ ionization mode. The total lipids were measured with a clinical kit (SPINREACT) as indicated by the manufacturer. This method is based in sulfo-phospho-vanillin reaction [18] .
2.5. Isolation and Analysis of Glycolipids
After seven days of growth, all cultures were collected and centrifuged at 2500 g at 4˚C. The media free of cells was lyophilized to dryness, weighed and kept at room temperature in desiccator until used. One fraction of the dried material (0.3 g) was dissolved in water. Ethyl acetate was added to a 1:1 ratio [1] . The organic phase was collected repeating the procedure to the remaining aqueous phase. The organic phase containing the glycolipids was rotary evaporated until we obtained a brownish sticky gum. A thin layer chromatography on silica plates (Silica-Fluorescent gel 60, Merck) was used to separate the glycolipids present using a mobile phase of chloroform-methanol-water (65:25:4 v/v/v). Detection of glycolipids was done with a UV lamp at short wavelength, or using a p-Anisaldehyde spray prepared by mixing 50 ml acetic acid, 1 ml of sulfuric acid and 0.5 ml of p-any- saldehide. HPLC separation of the glycolipids was performed with a HPLC LC-10ATVP Shimadzu apparatus and 10 mm, 4.6 mm × 30 cm C18 µ Bondapak column. A linear gradient was applied at a flow rate of 1 mL∙ min−1, from 0% to 100% of solvent B (acetonitrile) in 5 min and then holding 15 min at 100% solvent B. Eluted fractions were detected at 240 nm and analyzed for emulsifying capacity.
2.6. Emulsification Index Determination
The emulsification capacity was estimated according the emulsification index (E24) as described in [19] with the following modifications. One ml of kerosene was mixed (v/v) with 100 mg of the dried sample dissolved in 1 ml of water in a flat bottom test tube and shaken at high speed for 2 minutes. After 24 hours, the height of the stable emulsion layer was measured. The emulsion index E24 was calculated as the ratio of the height of the emulsion layer and the total height of the liquid. As negative and positive controls, we used H2O/kerosene and 1 mg∙mL−1 Surfactin/kerosene respectively. All data were expressed as percentages by the formula:
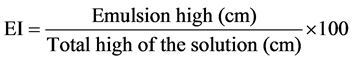
3. Results
The growth of U. maydis cells in different nitrogen media was followed for seven days and compared to a complete rich YPD media. The case of MM media with no added nitrogen was also considered (MM−N). Significant differences in the growth followed by optical density at 600 nm of the four cultures were observed when the nitrogen source was limiting or the cells were nitrogen-starved (Figure 1). Cells grown in YPD showed an exponential phase during the first 24 h of incubation followed by a stationary phase that was reached slowly at 108 - 120 h of the growth curve. A similar behavior was observed for cells grown in MM containing nitrate as nitrogen source: cells reached the stationary phase around 144 h of the culture with an initial exponential phase within the first 24 h of the growth. In the case of MM+U, the exponential growth rate was dilatory smaller than the rates found for the other culture conditions, and this was associated with a low yield as well as the stationary stage. Cells grown under nitrogen starvation (MM−N) show a short exponential phase followed by an early stationary phase; as expected, in this growth condition there was a smaller yield in biomass production. Biomass was calculated using the OD600 and translated into cells mL−1 (1 OD corresponds to 5 × 105 cells mL−1). The following values were obtained for the 5th day of culture: YPD 11.6 × 105, MM+N 8.7 × 105, and MM−N (or MM+U) 6.5 × 105. These results indicated that urea is a poor source of nitrogen for U. maydis and the stress produced in the fungus is similar to the one observed in the absence of nitrogen source.
Cells were observed by light microscopy during the period of culture. Figure 2 shows cells at the fifth day. Cells grown in YPD showed the characteristic cylindrical shape of U. maydis (Figure 2(A)). Yeasts cultured in MM with nitrate presented multiple transversal dark septa along the main axis of the cell (Figure 2(B)). In MM+U, cells showed irregular vesicles (Figure 2(C)) that increased in size when cells were incubated for longer times (data not shown).
Cells grown in glucose without nitrogen source showed round refractive bodies under light microscopy (Figure 2(D)). Those cells were stained with the molecular probe BODIPY 493/503 which is specific for cellular lipid droplets and stain-neutral lipids. Figure 3 shows cells cultured in MM−N (C, D) compared to YPD grown cells (A, B) viewed under normal and fluorescence microscopy (100× magnifications). There was a remarkable difference of accumulated neutral lipids in nitrogen starved and normal cells (Figure 3(B), Figure 3(D)). Discrete lipid deposits were more defined in starved cells than in YPD grown cells. The lipids were extracted and measured as total lipids.
Figure 1. U. maydis cells were grown in rich medium (YPD) and under either nitrogen limitation or starvation, with glucose as carbon source. All cultures were inoculated with 50 OD /L and shaked at 125 rpm for 7 days at 29˚C. Yeast peptone dextrose medium (YPD
Figure 2. U. maydis cells micrographics taken at the fifth day of growth. Light microscopy was performed at 100× with oil immersion objective. A) Cells in YPD Medium. B) Cells growing in Minimum media with nitrate (MM). C) Minimum media with urea. D) Cells grown in Minimum media without a nitrogen source (MM−N). All cells were cultured at 29˚C, 125 rpm during 7 days.
Figure 3. Light and fluorescence microscopy of U. maydis cells. (A)-(C) Cells cultured in YPD (upper panel) or MM-N (Lower panel) for 3 days were viewed under light microscopy. (B)-(D) Same preparations showing the accumulation of lipids (upper panel YPD; lower panel MM-N). For light microscopy a 100× oil immersion objective was used. For fluorescence microscopy, BODYPI 495/593 was used as a probe for lipid bodies.
The amount of lipids accumulated by cells were: MM−N with 0.381 mg∙mL−1, followed by YPD cells (0.26 mg∙mL−1), MM+N (0.19 mg∙mL−1) and MM+U with the lowest value (0.047 mg∙mL−1) (Figure 4). Composition of the fatty acids extracted from U. maydis grown in the four cultured media is presented in Table 1. Unsaturated fatty acids C18:1 and C24:3 from cells cultured in YPD were around 64%, in MM+N only C18:1 was detected in 54.9% and in cells deprived of nitrogen source, C16:1 and C18:1 overtake the 66.9%. The saturated lipids in all these cases corresponded to C16:0 and C18:0 in different proportions. Interestingly, in cells grown in MM+U the lipids were all saturated (99%). In all conditions a short fatty acid (C5:0) was present.
The glycolipids produced under nitrogen limitation or starvations were separated by TLC. As shown in Figure 5, very few UA molecules and one band of MEL were produced in MM+N with 0.3% of potassium nitrate as a
Figure 4. Total lipid concentration in cell extracts of U. maydis in the different media, taken at the fifth day of growth. Bars show the mean ± SD.
Table 1. Fatty acids (FA) detected by GC/MS.
Lipids were extracted with organic solvents from cells grown in each culture media as mentioned in methods.
Figure 5. Thin layer chromatography (TLC) of organic extract from cells cultured in different media. Minimum medium with nitrate (MM). Minimum medium without a nitrogen source (MM−N). Minimum medium with urea as a nitrogen source (MM+U).
nitrogen source. For cells grown in nitrogen-starved media (MM−N), high content of one class of UA molecules was identified as a lower band, and in MM+U a mix of UA in lower concentration and different bands of MEL were found.
The emulsification capacity of the glycolipids produced by nitrogen limitation or starved cells was evaluated with kerosene and compared to emulsification by surfactin. As shown in Figure 6, the emulsification capacity of the glycolipids produced in these three media gave an average of 72% with respect to the control.
The HPLC separation of glycolipids showed different distributions of hydrophobic compounds (Figure 7). Fractions were collected and analyzed in terms of their emulsifying capacity. Next, HPLC fractions containing glycolipids with emulsifying capacity were pooled.
For cells grown in MM+N, two main batches containing F4-5-6 and F10-11-12-13 were selected (Figure 8(A)). Glycolipid from MM−N was obtained just in one fraction (F7), although F14 also showed a small emulsifying capacity (Figure 8(B)). Surfactants produced during the growth in MM+U were pooled as F1-2, F5-6, and F7-8 (Figure 8(C)). The emulsifying capacity of all fractions pooled from HPLC purification still remained at the same average value of 72% as indicated in the bottom of Figure 8.
Figure 6. Emulsification capacity of glycolipids isolated from cells grown in different mimimal media. With nitrate (MM+N); without a nitrogen source (MM−N); with urea (MM+U). Emulsification capacity of the glycolipids extracted was compared to the capacity of Surfactin. All glycolipids from U. maydis gave an average emulsification of 72% - 76%.
Figure 7. Elution profiles of the organic extracts separated by reverse phase HPLC. (A) Minimum medium with nitrate. (B) Minimum medium without a nitrogen source. (C) Minimum medium with urea as nitrogen source. Eluted fractions were followed by 240 nm absorption.
Figure 8. Emulsification index of the fractions obtained by reverse phase HPLC. (A) Minimum medium with nitrate. (B) Minimum medium without a nitrogen source. (C) Minimum medium with urea as nitrogen source.
4. Discussion
U. maydis is a phytopathogen basidiomycete that grows at the expenses of the nutrients given by the host. Although it is not an oily yeast, U. maydis is able to grow on oily substrates such as free fatty acids, and produce glycolipids upon nitrogen limitation [9] [11] [20] [21] . Our study confirms that under nitrogen-limitation U. maydis produced glycolipids differentially and that nitrogen starvation increases the accumulation of neutral lipids favoring the production of a single type of glycolipid (UA) as reported in the study of Nit2 control which is involved in nitrogen catabolite repression [22] . Additionally, we demonstrated that the glycolipid and fatty acid profile differs in each culture media containing low nitrogen or absence of it. Lipid accumulation is influenced by the C/N ratio (optimum 100/1) as described for oily yeasts [15] [23] and U. maydis accumulates the highest amount of lipids when only glucose is present in the cultured media. Lipids as carbon source were not used in our study because the extraction of glycolipids and fatty acids is easier when a soluble carbon source is used, and our goal was to assess the effect of nitrogen source in a media without interference. Glucose is the most commonly employed carbon source, so the excess of carbon may be channeled into triacylglycerol (TAG) synthesis [24] . TAG accumulation is triggered by cells exhausting nitrogen from the culture medium, and in this condition, glucose continues to be assimilated [25] . In oily yeasts has been proposed that during the synthesis of fatty acid the glucose intake accelerate the citrate accumulation at the cytosol, after the conversion of pyruvate to Acetyl-CoA and the subsequent citrate formation by condensation with oxaloacetate in the tricarboxylic acid cycle in the mitochondria. Cytosolic ATP-Citrate lyase is necessary to recover the Acetyl CoA in the cytosol for use in lipid synthesis, and U. maydis must use this route due to the presence of an ATP-Citrate lyase gene (UM 01005.1) [26] [27] . During the production of itaconic acid (methylene succinic acid) in U. mayids during nitrogen-limited conditions, it has been suggested that the relative oxygen content decreased, making the biomass more reduced and allowing for the accumulation of less oxygenated compounds as hydrocabons [28] . These authors also included in their study the influence of nitrogen supply and found that nitrogen limitation increased the cell size, probably due to the accumulation of lipids.
All eukaryotic organisms and some gram-positive bacteria store triacylglycerols in intracellular compartments which are known as lipid particles, lipid droplets (LDs), lipid bodies (LBs), oil bodies, oleosomes, or spherosomes (in plants) [29] . We observed that U. maydis accumulated neutral lipids when it is deprived of nitrogen source in contrast to cells cultured in the presence of this nutrient. Lipid bodies have been described in other organism as yeast and algae [30] [31] . The accumulation of lipids in nitrogen starved cells might be explained by authophagy, a mechanism used by organisms in response to change in nutrient supply and allows the cells to alter LB metabolism to meet cellular energy demand [25] [32] . Accumulated fatty acids were principally unsaturated, as determined by the vainillin method, and only urea cultures showed a high amount of saturated lipids. Those results disagree with the information about UA and MEL lipid moieties. The longest lipid moiety of ustilagic acid has been reported to hydroxilated C16:0 but for mannosylerytritol lipids C14, C16 and C18 saturated and unsaturated fatty acids were found although sucrose was the carbon source [8] [33] . The enzymes involved in the biosynthesis of the glycolipids (UA and MEL) have been described in [11] [12] . For UA a cytochrome P450 monooxygenase Cyp1 catalyzes terminal hydroxylation of palmitic acid and cyp2 is required for subterminal hydroxylation (w-1 hydroxylation) of this fatty acid [1] [12] . The expression of cyp1 is not under nitrogen regulation [22] which implies that UA production can occur without nitrogen source. It has also been reported that the fatty acid profile of MEL contains C14:0, C16:0, C14:1 and C16:1 in U. nuda, but the enzymes involved in the synthesis of this molecule have not been identified [34] . If fatty acids are accumulated they can be the raw material to be processed and join to carbohydrate molecules involving enzymes as lipases [35] . So biosurfactant secreted into the culture medium containing lipids and carbo hydrate moieties involve a large amount of enzyme synthesis, but do not yet know how this happens. In particular TAGs accumulation intracellular can be confused with glycolipids because both contain fatty acids and some stain such as nile red can give the same reaction [36] . In this sense, glycolipids cannot be detected by conventional techniques, and is only by detection of the lipid or sugar moiety that they can be measured. Due to this problem, we used BODIPY to identify accumulation of TAGs in cells grown in MM-N, in spite of the low yield of biomass due to the triglycerides inside the cells without interference of other lipids, allowing the lack of nitrogen.
In agreement with other reports, the preference of these two glycolipids is affected considerably by the nitrogen source [1] [37] . Additionally, in this study we showed first that UA is the main glycolipid when esterified fatty acids are accumulated in the lipid bodies, and second, that U. maydis is an important cell factory that produces not only glycolipids, but also fatty acids which may have potential application in biofuels.
5. Conclusions
1) The presence or absence of nitrogen source affected the morphology and lipid metabolism of U. maydis yeast.
2) Cells grown in media containing glucose as carbon source but without any nitrogen source accumulated unsaturated fatty acids in lipid bodies.
3) Glycolipid selectivity was under nitrogen source influence.
Acknowledgements
The authors thank to Dr. Luis Vasquez Mendez for his help in microscopic analysis and Eddie Medina from Shimadzu Scientific Instruments Analytical Latin America Business Manager for the use of the HPLC equipment. Thanks are given to Dr Monica Montero Lomelí and Bruno Bozaquel Morais Universidad Federal Do Rio de Janeiro Brasil for the valuable help with fluorecense stain and helpful discussion.
This work was supported by the following grants: Consejo de Ciencia y Tecnología D.F (ICyT D.F PIFUTP08-131), Instituto Politécnico Nacional (SIP 20101135-20110773-20120823), Universidad Nacional Autónoma de México (PAPIIT IN201511, IN209614) and CONACyT 59855, 166472.
Ariana Zavala-Moreno was supported by CONACYT scholarship (174657) and SIP-PIFI. IPN.
References
- Hewald, S., Josephs, K. and Bölker, M. (2005) Genetic Analysis of Biosurfactant Production in Ustilago maydis. Applied and Environmental Microbiology, 71, 3033-3040. http://dx.doi.org/10.1128/AEM.71.6.3033-3040.2005
- Brefort, T., Doehlemann, G., Mendoza-Mendoza, A., Reissman, S., Djamei, A. and Khaman, R. (2009) Ustilago maydis as a Pathogen. Annual Review of Phytopathology, 47, 423-445. http://dx.doi.org/10.1146/annurev-phyto-080508-081923
- Spoeckner, S., Wray, V., Nimtz, M. and Lang, S. (1999) Glycolipids of the Smut Fungus Ustilago maydis from Cultivation on Renewable Resources. Applied Microbiology and Biotechnology, 51, 33-39. http://dx.doi.org/10.1007/s002530051359
- Morita, T., Kitagawa, M., Suzuki, M., Yamamoto, S., Sogabe, A., Yanagidani, S., Imura, T., Fukuoka, T. and Kitamoto, D. (2009) A Yeast Glycolipid Biosurfactant, Mannosylerythritol Lipid, Shows Potential Moisturizing Activity toward Cultured Human Skin Cells: The Recovery Effect of MEL-A on the SDS-Damaged Human Skin Cells. Journal of Oleo Science, 58, 639-642. http://dx.doi.org/10.5650/jos.58.639
- Morita, T., Fukuoka, T., Imura, T. and Kitamoto, D. (2013) Production of Mannosylerythritol Lipids and Their Application in Cosmetics. Applied Microbiology and Biotechnology, 97, 4691-4700. http://dx.doi.org/10.1007/s00253-013-4858-1
- Kitamoto, D., Isoda, H. and Nakahara, T. (2002) Functions and Potential Applications of Glycolipid Biosurfactants from Energy-Saving Materials to Gene Delivery Carriers. Journal of Bioscience and Bioengineering, 94, 187-201. http://dx.doi.org/10.1016/S1389-1723(02)80149-9
- Makkar, R.S. and Cameotra, S.S. (2002) An Update on the Use of Unconventional Substrates for Biosurfactant Production and Their New Applications. Applied Microbiology and Biotechnology, 58, 428-434. http://dx.doi.org/10.1007/s00253-001-0924-1
- Morita, T., Ishibashi, Y., Fukuoka, T., Imura, T., Sakai, H., Abe, M. and Kitamoto, D. (2009) Production of Glycolipid Biosurfactants, Mannosylerythritol Lipids, Using Sucrose by Fungal and Yeast Strains, and their Interfacial Properties. Bioscience, Biotechnology, and Biochemistry, 73, 2352-2355. http://dx.doi.org/10.1271/bbb.90439
- Liu, Y., Koh, C.M. and Ji, L. (2011) Bioconversion of Crude Glycerol to Glycolipids in Ustilago maydis. Bioresource Technology, 102, 3927-3933. http://dx.doi.org/10.1016/j.biortech.2010.11.115
- Amaral, P.F., Coelho, M.A., Marrucho, I.M. and Coutinho, J.A. (2008) Biosurfactants from Yeasts: Characteristics, Production and Application. Advances in Experimental Medicine and Biology, 627, 236-249.
- Hewald, S., Linne, U., Scherer, M., Marahiel, M.A., Kämper, J. and Bölker, M. (2006) Identification of a Gene Cluster for Biosynthesis of Mannosylerythritol Lipids in the Basidiomycetous Fungus Ustilago maydis. Applied and Environmental Microbiology, 72, 5469-5477. http://dx.doi.org/10.1128/AEM.00506-06
- Teichmann, B., Linne, U., Hewald, S., Marahiel, M.A. and Bölker, M. (2007) A Biosynthetic Gene Cluster for a Secreted Cellobiose Lipid with Antifungal Activity from Ustilago maydis. Molecular Microbiology, 66, 525-533. http://dx.doi.org/10.1111/j.1365-2958.2007.05941.x
- Guo, Y., Cordes, K.R., Farese Jr., R.V. and Walther, T.C. (2009) Lipid Droplets at a Glance. Journal of Cell Science, 122, 749-752. http://dx.doi.org/10.1242/jcs.037630
- Liu, Y.M., Zhang, C.Y., Shen, X.P., Zhang, X.L., Cichello, S., Guan, H.B. and Liu, P.S. (2013) Microorganism Lipid Droplets and Biofuel Development. BMB Reports, 46, 575-581. http://dx.doi.org/10.5483/BMBRep.2013.46.12.271
- Finn, P.F. and Dice, J.F. (2006) Proteolytic and Lipolytic Response to Starvation. Nutrition, 22, 830-844. http://dx.doi.org/10.1016/j.nut.2006.04.008
- Holliday, R. (1961) The Genetics of Ustilago maydis. Genetical Research, 2, 204-230. http://dx.doi.org/10.1017/S0016672300000719
- Bozaquel-Morais, B.L., Madeira, J.B., Maya-Monteiro, C.M., Masuda, C.A. and Montero-Lomeli, M. (2010) A New Fluorescence-Based Method Identifies Protein Phosphatases Regulating Lipid Droplet Metabolism. PLoS ONE, 5, e13692. http://dx.doi.org/10.1371/journal.pone.0013692
- Knight, J.A., Anderson, S. and Rawle, J.M. (1972) Chemical Basis of the Sulfo-Phospho-Vanillin Reaction for Estimating Total Serum Lipids. Clinical Chemistry, 18, 199-202.
- Techaoei, S., Leelapornpisid, P., Santiarwarn, D. and Lumyong, S. (2007) Preliminary Screening of Biosurfactant- Producing Microorganisms Isolated from Hot Spring and Garages in Northern Thailand. KMITL Science and Technology Journal, 7, 38-43.
- Bölker, M., Basse, C.W. and Schirawsk, J. (2008) Ustilago maydis Secondary Metabolism—From Genomics to Biochemistry. Fungal Genetics and Biology, 45, S88-S93. http://dx.doi.org/10.1016/j.fgb.2008.05.007
- Morita, T., Fukuoka, T., Imura, T. and Kitamoto, D. (2013) Accumulation of Cellobiose Lipids under Nitrogen-Limiting Conditions by Two Ustilaginomycetous Yeasts, Pseudozyma aphidis and Pseudozyma hubeiensis. FEMS Yeast Research, 13, 44-49. http://dx.doi.org/10.1111/1567-1364.12005
- Horts, R.J., Zeh, C., Saur, A., Sonnewald, S., Sonnewald, U. and Voll, L.M. (2012) The Ustilago maydis Nit2 Homolog Regulates Nitrogen Utilization and Is Required for Efficient Induction of Filamentous Growth. Eukaryotic Cell, 11, 368-380. http://dx.doi.org/10.1128/EC.05191-11
- Ageitos, J.M., Vallejo, J.A., Veiga-Crespo, P. and Villa, T.G. (2011) Oily Yeasts as Oleaginous Cell Factories. Applied Microbiology and Biotechnology, 90, 1219-1227. http://dx.doi.org/10.1007/s00253-011-3200-z
- Hsieh, H.J., Su, C.H. and Chien, L.J. (2012) Accumulation of Lipid Production in Chlorella minutissima by Triacylglycerol Biosynthesis-Related Genes Cloned from Saccharomyces cerevisiae and Yarrowia lipolytica. Journal of Microbiology, 50, 526-534. http://dx.doi.org/10.1007/s12275-012-2041-5
- Beopoulos, A., Chardot, T. and Nicaud, J.M. (2009) Yarrowia lipolytica: A Model and a Tool to Understand the Mechanisms Implicated in Lipid Accumulation. Biochimie, 91, 692-696. http://dx.doi.org/10.1016/j.biochi.2009.02.004
- Hynes, M.J. and Murray, S.L. (2010) ATP-Citrate Lyase Is Required for Production of Cytosolic Acetyl Coenzyme A and Development in Aspergillus nidulans. Eukaryotic Cell, 9, 1039-1048. http://dx.doi.org/10.1128/EC.00080-10
- Liu, Z.J., Gao, Y., Chen, J., Imanaka, T., Bao, J. and Hua, Q. (2013) Analysis of Metabolic Fluxes for Better Understanding of Mechanisms Related to Lipid Accumulation in Oleaginous Yeast Trichosporon cutaneum. Bioresource Technology, 130, 144-151. http://dx.doi.org/10.1016/j.biortech.2012.12.072
- Klement, T., Milker, S., Jäger, G., Grande, P.M., Domínguez de María, P. and Büchs, J. (2012) Biomass Pretreatment Affects Ustilago maydis in Producing Itaconic Acid. Microbial Cell Factories, 11, 43. http://dx.doi.org/10.1186/1475-2859-11-43
- Walther, T.C. and Farese Jr., R.V. (2009) The Life of Lipid Droplets. Biochimica et Biophysica Acta, 1791, 459-466. http://dx.doi.org/10.1016/j.bbalip.2008.10.009
- Morita, E., Kumon, Y., Nakahara, T., Kagiwada, S. and Noguchi, T. (2010) Docosahexaenoic Acid Production and Lipid- Body Formation in Schizochytrium limacinum SR21. Marine Biotechnology, 8, 319-327. http://dx.doi.org/10.1007/s10126-005-5060-y
- Siaut, M., Cuiné, S., Cagnon, C., Fessler, B., Nguyen, M., Carrier, P., Beyly, A., Beisson, F., Triantaphylidès, C., Li-Beisson, Y. and Peltier, G. (2011) Oil Accumulation in the Model Green Alga Chlamydomonas reinhardtii: Characterization, Variability between Common Laboratory Strains and Relationship with Starch Reserves. BMC Biotechnology, 11, 7. http://dx.doi.org/10.1186/1472-6750-11-7
- Levine, B. and Klionsky, D.J. (2004) Development by Self-Digestion: Molecular Mechanisms and Biological Functions of Autophagy. Developmental Cell, 6, 463-477. http://dx.doi.org/10.1016/S1534-5807(04)00099-1
- Lemieux, R.U. and Charanduk, R. (1951) Biochemistry of the Ustilaginales VI. The Acyl Groups of Ustilagic Acid. Canadian Journal of Chemistry, 29, 759-766. http://dx.doi.org/10.1139/v51-087
- Bhattacharjee, S.S., Haskins, R.H. and Gorin, P.A.J. (1970) Location of Acyl Groups on Two Acylated Glycolipids from Strains of Ustilago (Smut Fungi). Carbohydrate Research, 13, 235-246. http://dx.doi.org/10.1016/S0008-6215(00)80830-7
- Buerth, C., Kovacic, F., Stock, J., Terfüchete, M., Wilhelm, S., Jaeger, K.E., Feldbrügge, M., Schipper, K., Ernest, J.F. and Tielker, D. (2014) Uml2 Is a Novel CalB-Type Lipase of Ustilago maydis with Phospholipase A Activity. Applied Microbiology and Biotechnology, 98, 4963-4973. http://dx.doi.org/10.1007/s00253-013-5493-6
- Morita, T., Konishi, M., Fukuoka, T., Imura, T. and Kitamoto, D. (2007) Physiological Differences in the Formation of the Glycolipid Biosurfactants, Mannosylerythritol Lipids, between Pseudozyma antarctica and Pseudozyma aphidis. Applied Microbiology and Biotechnology, 74, 307-315. http://dx.doi.org/10.1007/s00253-006-0672-3
- Irudayaraj, J., Bhaduri, S., Uppara, P.V. and Doble, M. (2008) Mannosylerythritol Lipids: A Review. Journal of Industrial Microbiology & Biotechnology, 35, 1559-1570. http://dx.doi.org/10.1007/s10295-008-0460-4
Abbreviations and Acronyms
UA: Ustilagic acid; MEL: Mannosylerythritol lipid; YPD: Yeast, peptone, dextrose; LD: Lipid droplets; LB: Lipid bodies; MM+N: Minimal media with nitrogen source; MM−N: Minimal media without nitrogen source; MM+U: Minimal media with urea like nitrogen source; TAG: Triacylglycerol; FA: Fatty acid.
NOTES

*Corresponding author.










