Journal of Cancer Therapy
Vol.2 No.1(2011), Article ID:4103,8 pages DOI:10.4236/jct.2011.21001
Comparative Proteomics Analysis of Exhaled Breath Condensate in Lung Cancer Patients
![]()
1Inflammation and Infection Research Centre and Prince of Wales Clinical School, Faculty of Medicine, University of New South Wales, Sydney, Australia;
2Department of Respiratory Medicine, Prince of Wales Hospital, Randwick, Australia;
3Department of Medical Oncology, Prince of Wales Hospital, Randwick, Australia;
4Bioanalytical Mass Spectrometry Facility, University of New South Wales, Sydney, Australia.
Email: paul.thomas@unsw.edu.au
Received November 29th, 2010; revised February 20th, 2011; accepted February 28th, 2011.
Keywords: Lung Cancer, Exhaled Breath Condensate, Mass Spectrometry, Proteomics
ABSTRACT
The prognosis for patients with non-small cell lung cancer (NSCLC) remains poor in spite of better treatments. This relates mainly to the fact that the majority of patients present with advanced disease. There is a need to identify tools which can improve screening for lung cancer in the at risk patient population. The aim of this study was to compare the breath proteomic profile of NSCLC patients with healthy control subjects to explore the potential of new biomarkers of lung cancer. Comparative proteomic analysis of exhaled breath condensate (EBC) between 14 patients with NSCLC and 13 healthy control subjects were carried out using LTQ FT Ultra mass spectrometry and database searching to determine any unique proteins. In total, 29 unique proteins were identified using multiple protein identification algorithms. A comparison of lung cancer, smoker, and ex-smoker proteomes showed that 18 proteins were shared among the three groups. While one unique protein was found in smokers and lung cancer patients, four proteins were unique to ex-smokers. This data set provides a foundation for evaluation of these proteins from EBC as potential biomarkers for non-invasive lung cancer diagnosis.
1. Introduction
Lung cancer is a major cause of cancer-related death in industrialized countries worldwide [1]. Non-small cell lung cancer (NSCLC) accounts for about 80% of all lung cancers. Despite advances in diagnosis and treatment strategies in the last decade, prognosis of NSCLC patients is poor with a 5-year overall survival of 16%, the lowest of any cancer [2,3]. Two major reasons account for this poor clinical picture. Firstly, carcinogenesis of lung cancer is a complex process involving multiple events and steps. Although molecular pathogenesis studies on lung cancer have been undertaken successfully on the gene (DNA), transcription (mRNA) and protein levels, the carcinogenic mechanism remains unclear [4]. Secondly, although outcomes have improved in the treatment of NSCLC, including better chemotherapy and targeted agents [5] more than 75% of patients will have advanced or metastatic disease at presentation, due to lack of early symptoms which might be improved by screening or early diagnostic tools [6]. If lung cancer is localised at the time of diagnosis and treated promptly by surgery, the 5-year survival rate improves to 52% [7]. The benefits of early diagnosis have prompted research into screening methods for early stage lung cancer in high-risk or smoking populations. Diagnosis of lung cancer is performed currently by sputum cytology, interval chest x-rays, computed tomography (CT) scans, and bronchoscopy, with cytopathologic examination of bronchoalveolar lavage, endobronchial brushings and biopsies obtained from the area of abnormality. Most of these tests are invasive and expensive, and there are often delays in establishing the diagnosis and initiating treatment. To improve lung cancer management and survival, there is a need to develop screening and early diagnostic strategies that are sensitive, specific, and non-invasive.
In terms of disease diagnosis and prognosis, exhaled breath condensate (EBC) appears to be an attractive method that offers several advantages. It is a simple, safe, inexpensive, and non-invasive method of sampling the lower respiratory tract in humans, particularly as it directly samples the local lung environment [8]. The condensate contains a range of nonvolatile substances [9] including macromolecules such as proteins [10,11]. Specific proteins have been identified in EBC that may act as significant markers of lung cancer. For example; endothelin-1, tumour necrosis factor-α, and interleukin-6 were found to be elevated in the EBC of NSCLC patients when compared to healthy controls [12,13], however, these markers have limited sensitivity and specificity for early diagnosis.
Proteomics has recently been introduced to the field of cancer research, and its potential applications are just beginning to be understood. Unlike the study of a single protein, proteomic technology offers a systemic overview that provides the potential to improve our understanding of lung cancer. Proteomic analysis of biomarkers of lung cancers in human plasma, serum and tissues have to date utilised a wide variety of technologies such as the use of two-dimensional gel electrophoresis (2D-PAGE) for separation of tumor proteins with protein identification using matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS), as well as the alternate technology of surface-enhanced laser desorption ionization mass spectrometry (SELDI-MS) [8]. Subsequently, the molecular basis for the development and progression of the disease can be investigated. The normal serum proteome provides the background against which the biomarkers representing “abnormality” can be identified.
Changes in the protein profile secreted into the lower respiratory tract may be detected in EBC of lung cancer patients as an indication of the underlying neoplastic processes, presenting a possible screening tool for the early detection of lung carcinomas [8]. EBC is a challenging subject for the study of proteomics because of its complexity and low levels of protein. Biomarker proteins are often present in small amounts and detection of low-abundance protein is dependent on the complexity of the protein mixture in addition to the power of resolution and sensitivity of the separation and identification method employed. An alternate approach is direct analysis of a complex protein mixture without protein separation using LTQ FT Ultra Mass Spectrometer. This system is a fully integrated hydrid mass spectrometery consisting of a Linear Ion Trap Mass Spectrometer, LTQ XL, combined with a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. The sensitivity of this machine is in the attomole range for peptides.
In an effort to further our understanding of lung cancer biology and to identify new candidate biomarkers to be used in the management of lung cancer, the aim of this study was to probe EBC with the LTQ FT Ultra Mass Spectrometry to conduct a proteomic comparison between patients with NSCLC and matched volunteers.
2. Materials and Methods
2.1. Subjects
The study was approved by the South Eastern Sydney & Illawarra Area Health Service Research Ethics Committee. Subjects were recruited from the Prince of Wales Hospital from July 2008 to July 2009. Patients with newly diagnosed lung cancer were recruited from the respiratory and oncology clinics prior to treatment. The control group included subjects without lung cancer and no history of COPD or other respiratory conditions; matched for age, gender, and socioeconomic group. This group comprised smokers and ex-smokers, defined as not having smoked for at least 1 year. Informed consent was obtained prior to sample collection and a questionnaire conducted to obtain demographic details including past medical history, medications, smoking history as well as staging and histological typing of lung cancer as appropriate. Lung function was measured using a spirometer (Minato Autospiro AS-600; Minato Medical Science, Osaka, Japan).
2.2. Sample Collection
EBC was obtained from subjects breathing into a siliconised glass collection device, cooled by ice to 4˚C as previously validated [14]. This method has been shown to prevent salivary contamination. Subjects breathed through a disposable mouthpiece and a one-way non-rebreathing valve with a curved tube, leading into the cooling device to collect EBC which also served as a saliva trap. Subjects breathed at a normal frequency and tidal volume for a 15 minute period and maintained a dry mouth by periodically swallowing excess saliva. The samples were then transferred to a 1.5 mL protein LoBind Tube, de-aerated by argon degassing for 1 minute and immediately stored at –80˚C for subsequent analysis.
2.3. Protein Content of Exhaled Breath Condensate
Total protein concentration in each sample was measured by the bicinchoninic acid (BCA) assay using the QuantiPro BCA Assay Kit according to the manufacturer’s instructions (Sigma-Aldrich, Sydney, Australia). The protein levels are indicated through a colour scale of green (low levels) to purple (high levels). The samples were then measured by colorimetry for absorbance readings.
2.4. Protein Concentrate
To equalise and concentrate the protein in EBC, a simple ultrafiltration step was performed using a Microcon® YM-3 centrifugal filter devices (Millipore Crop., Sydney, Australia), following the manufacturer’s instructions.
2.5. Mass Spectrometry and Database Search
2.5.1. Trypsin Digestion of the Protein Samples in Solution
Samples as a protein solution (0.5 mg/25 µl) were incubated with 5 μL of 25 mM NH4HCO3 containing 10 ng/mL of trypsin (Promega, Annandale, NSW, Australia) for 14 hours at 37˚C. The samples were resuspended in water containing 0.05% heptafluorobutyric acid and 1% formic acid.
2.5.2. LTQ-FT Ultra Analysis
Mass spectrometry analyses were carried out as described previously [15] . Digest peptides were separated by nano-liquid chromatography using an Ultimate 3000 HPLC and autosampler system (Dionex, Amsterdam, Netherlands). Samples (2.5 µl) were concentrated and desalted onto a micro C18 precolumn (500 µm × 2 mm, Michrom Bioresources, Auburn, CA) with H2O:CH3CN (98:2, 0.05% HFBA) at 20 µl/min. After a 4 minute wash the pre-column was switched (Valco 10 port valve, Dionex) into line with a fritless nano column (75 µ × ~10 cm) containing C18 media (5 µ, 200 Å Magic, Michrom) manufactured according to Gatlin [16]. Peptides were eluted using a linear gradient of H2O:CH3CN (98:2, 0.1% formic acid) to H2O:CH3CN (64:36, 0.1% formic acid) at 350 nl/min over 30 minutes. High voltage (1800 V) was applied to a low volume tee (Upchurch Scientific, Oak Harbor, WA) and the column tip positioned ~0.5 cm from the heated capillary (T = 200˚C) of a LTQ FT Ultra (Thermo Electron, Bremen, Germany) mass spectrometer. Positive ions were generated by electrospray and the LTQ FT Ultra operated in data dependent acquisition mode (DDA). A survey scan m/z 350 - 1750 was acquired in the FT ICR cell (resolution = 100,000 at m/z 400, with an initial accumulation target value of 1,000,000 ions in the linear ion trap). Up to the 7 most abundant ions (>2500 counts) with charge states of +2 or +3 were sequentially isolated and fragmented within the linear ion trap using collisionally induced dissociation with an activation q = 0.25 and activation time of 30 ms at a target value of 30,000 ions. M/z ratios selected for MS/MS were dynamically excluded for 30 seconds.
2.5.3. Database Searches
Peak lists were generated using Mascot Daemon/extract_ msn (Matrix Science, London, England, Thermo) using the default parameters, and submitted to the database search program Mascot (version 2.1, Matrix Science). Search parameters were: precursor tolerance 4 ppm and product ion tolerances ± 0.6 Da; Met(O) specified as variable modification, enzyme specificity was trypsin, 1 missed cleavage was possible and the non-redundant IPI_human protein database searched (May 2009).
2.5.4. Criteria for Protein Identification
The Mascot search results from each sample were then loaded into Scaffold (version 2.01 Proteome Software Portland, OR), searched using X! Tandem and grouped into a single result based on disease. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 1 identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
2.6. Statistical Analysis
Statistical analyses were applied to compare the study group of lung cancer patients with control groups: ex-smokers and smokers. Patients were matched for age, gender and smo king status. All the data were analysed using GraphPad Prism® software Version 5.00 and SPSS 10.0 for Windows. If the distribution conformed to the Normal distribution, parametric tests were performed; if the distribution was not Normal, non-parametric tests were performed. A p-value of less than 0.05 was regarded as statistically significant.
3. Results
3.1. A Total of 27 Subjects Were Recruited
14 NSCLC patients, and 13 controls (eight ex-smokers and five smokers). Subject characteristics are summarized in Table 1.
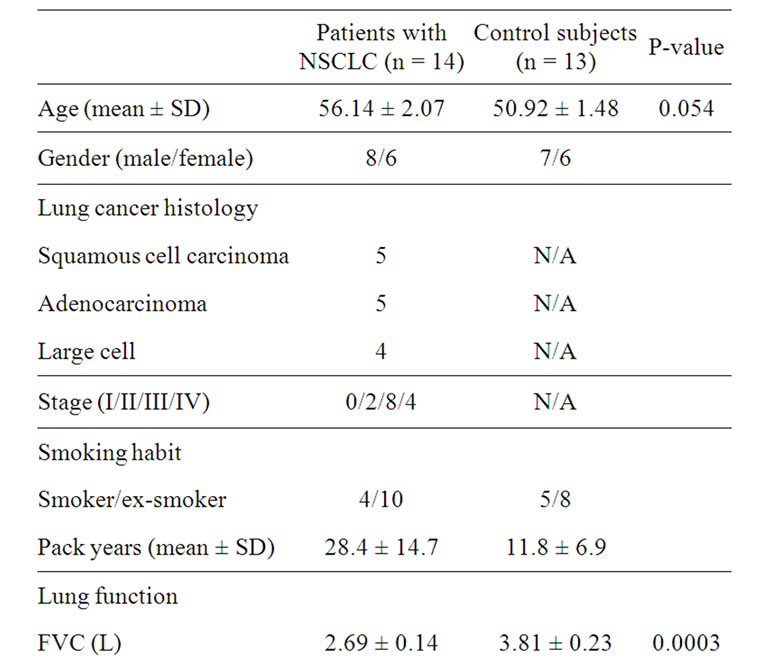
Table 1. Clinic pathological characteristics of patients with non-small cell lung cancer and control subjects.
3.2. Total Protein
The total protein level in each EBC sample available was measured using the micro BCA assay (Figure 1). No significant difference in total protein concentration was seen within the subgroups of control subjects (ex-smokers and current smokers) (p > 0.5 ANOVA). However, the concentration of total protein was found to be significantly increased in EBC of patients with lung cancer as compared to controls (4.204 ± 0.859 and 2.175 ± 0.3725 mg/µL, respectively, p < 0.05 ANOVA).
3.3. Protein Analysis
In total, 29 unique proteins were identified using multiple protein identification algorithms. For each protein identified, detailed identification data including accession number, molecular mass, and protein differences between normal and disease groups are shown in Table 2. Proteins with a Fisher exact p < 0.05 comparing lung cancer and control groups are shown. Keratin proteins are the major functional categories represented in the EBC, such as keratin 1, keratin 2, keratin 5, keratin 6A, keratin 9, and keratin 10. Within the 29 proteins, there were 4 proteins including keratin 14, keratin 16, a protein similar to hormerin, and keratin 17, which expressed significant differences between the lung cancer group and normal controls.
The comparison of the three groups (lung cancer, smoker, ex-smoker) is shown in Figure 2. A total of 29 proteins were detected in 27 samples based on the criteria as described in the methods. Of these, 21 proteins were detected in smokers, 26 proteins were detected in at least
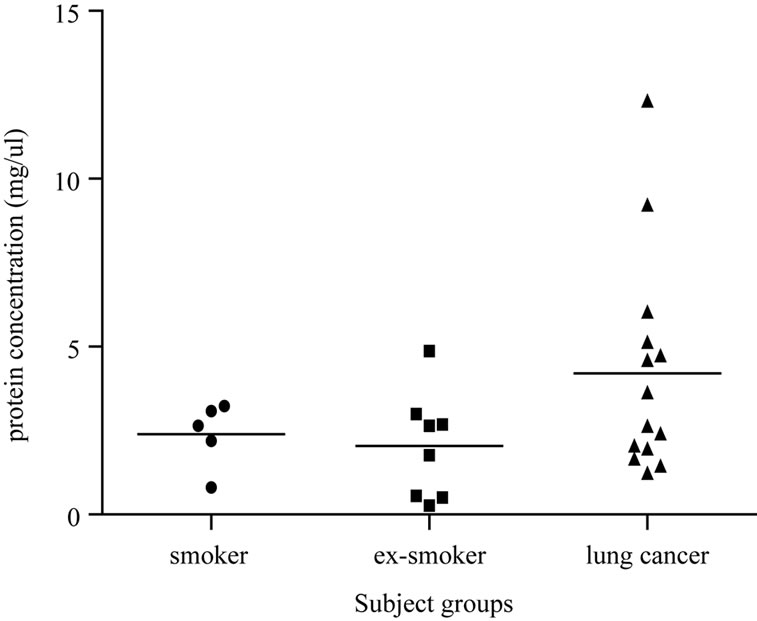
Figure 1. The graph shows the total protein concentrations measured in samples from ex-smokers, smokers and lung cancer patients. An analysis of the data produced a statistically significant difference observed across lung cancer patients compared to normal controls (p < 0.05).
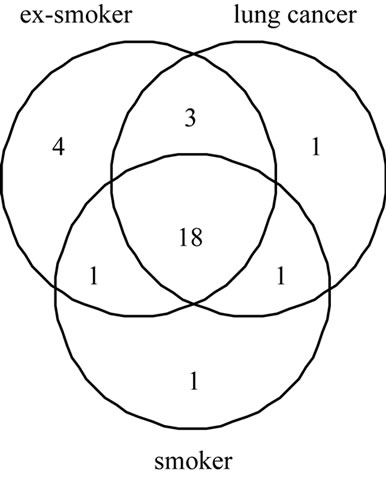
Figure 2. Venn diagram describing the proportion of proteins detected in ex-smokers, smokers and lung cancer. The circles represent proteins detected in at least one sample.
one ex-smoker and 23 proteins were detected in the majority of lung cancer patients. There were 18 proteins that were common to all three groups, while 1, 4, and 1 unique protein occurs in smokers, ex-smokers and lung cancer groups, respectively. In the lung cancer group the unique protein is growth hormone regulated TBC protein 1 (GRTP 1) seen in one patient.
4. Discussion
The proteome of EBC has the potential to be a valuable source of biomarkers for the diagnosis and therapeutic intervention of disease. Investigation of the differential proteome profile between healthy and lung cancer subjects can be a critical step in identifying possible diagnostic and prognostic biomarkers. EBC is a challenging subject for proteomic study because of its complexity and low range of protein concentration. Biomarker proteins are often present in small amounts and the detection of a low-abundance protein is dependent on the resolving power of the separation and identification method used. With respect to our study, several important observations about the presence of these proteins should be underlined. Firstly, refinements in EBC collection technique may improve the concentration of protein. In comparison to a simple glass device, siliconised glass and R-Tube systems, the EcoScreen system is able to collect larger amounts of total protein; however, the glass device is more portable and suitable to be taken to subjects who are unwell. All EBC collection systems tend to collect biomarkers which are at the lower limit of detection of specific assays. Siliconising the glass collection device may increase the yield of protein [14] but samples may still require concentration before measurement. Concentration by lyophilisation or filtration with desalting prior to analysis may be necessary to improve sensitivity and reproducibility [17], but the salts and other low molecular weight material will also be concentrated and this will
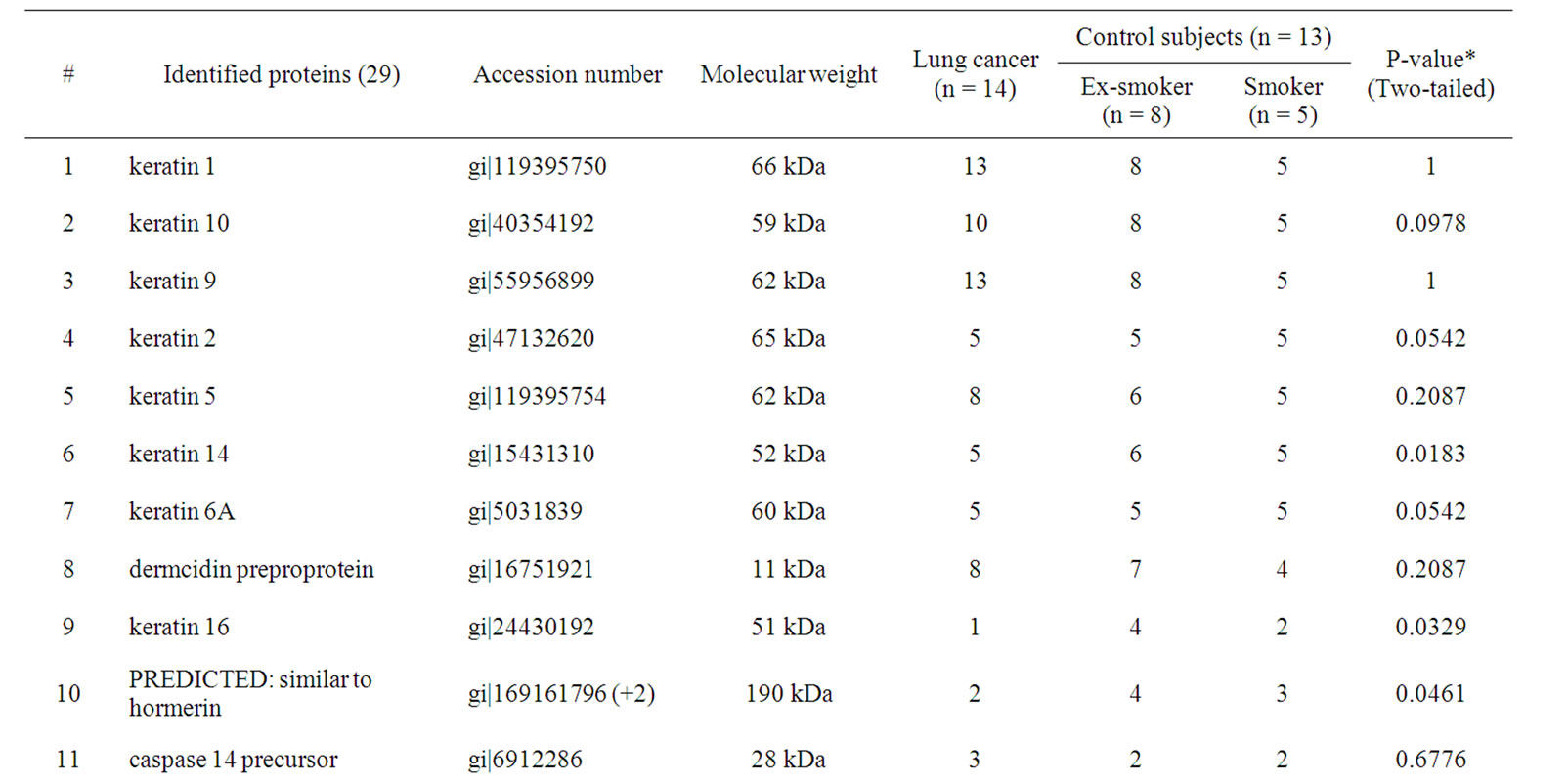
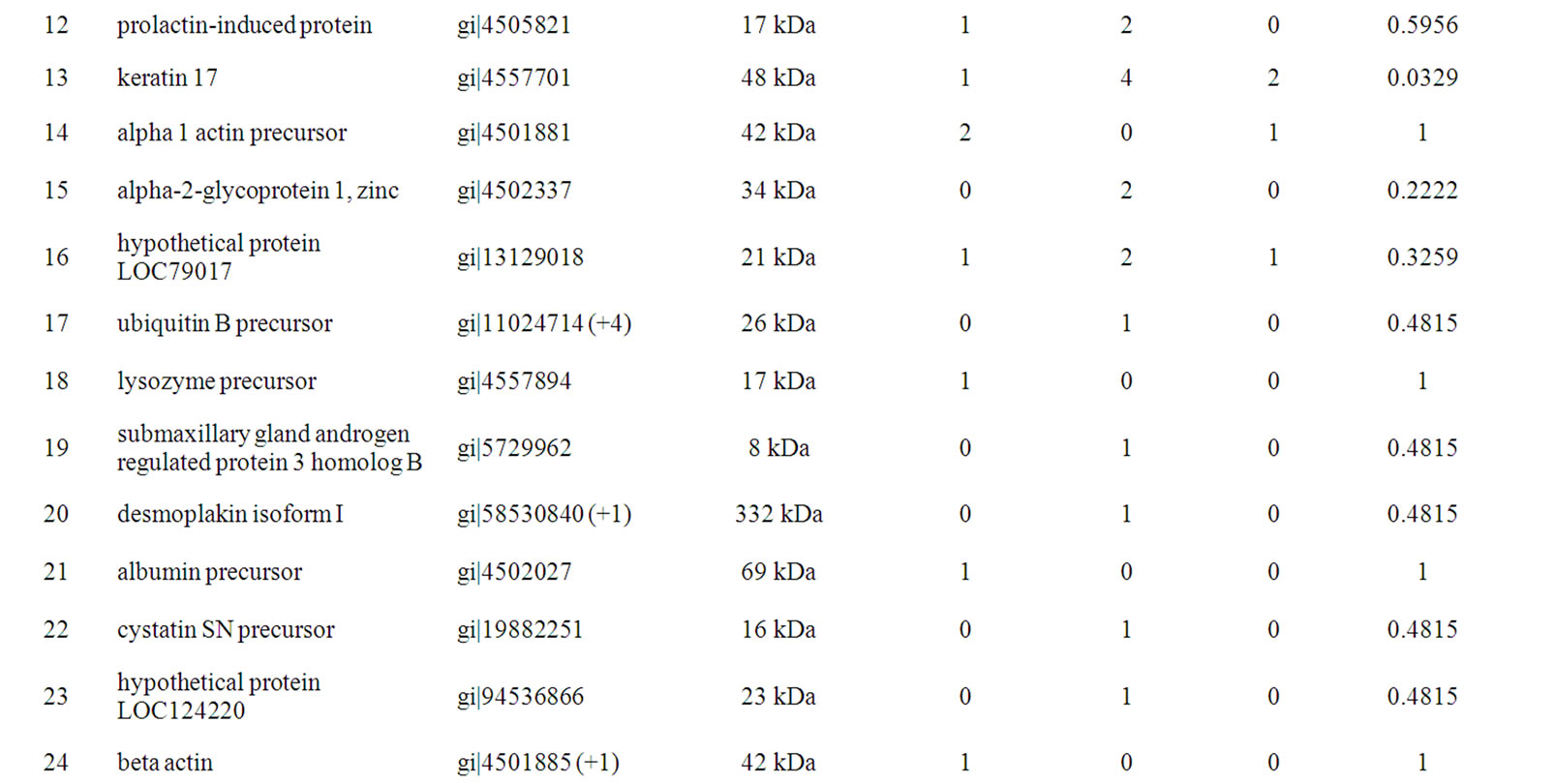

Table 2. Twenty-nine unique proteins were identified in EBC differed between lung cancer and control subjects.
reduce sensitivity or possibly contaminate mass spectrometry. Thus, using an ultrafiltration device to enrich the proteins appears to be the best option. Of note, the total protein levels were increased in the patients with lung cancer. While this did not obviously reflect elevated levels of a single protein this cannot be excluded by a qualitative method such as MS, but possibly it may represent either excess airway secretion or vascular leakage [18].
In proteomic studies, the Mowse cut-off scores for protein identification are very important. Higher Mowse scores reduce the number of identified proteins but the estimated false positive rate falls [19]. In this study, the cut-off value for protein identification was 95%. If a lower cut-off value was adopted (e.g. 80%), 33 proteins were identified and the results were almost the same as with the 95% value described. If the cut-off scores were 50%, we identified 69 proteins, but the estimated false positive rate was higher (results not shown). In total, 29 unique proteins were identified with high confidence using multiple protein identification in all the patients. Proteins such as keratin 1, keratin 2, keratin 5, keratin 6A, keratin 9, and keratin 10 were detected in almost all of the samples from the subjects investigated. Keratins have been observed as the major components in EBC samples [20]. EBC of smokers contain elevated amounts of CK9 and CK10 [21]. It is still largely a matter of speculation whether these molecules may be considered clinically relevant endpoints of pulmonary damage. Given the wide variation of results obtained in different studies, the data available in the literature still remain unclear. A few reports [21,22] support the notion that EBC cytokeratins may be a tool for monitoring lung inflammation, although others suggest that some EBC protein content may be derived from ambient air but this is unlikely to represent a significant issue. It is possible that EBC is contaminated by keratin 1 derived from human skin but it is quite likely to be derived from the upper airway [23].
An interesting observation from our study was the finding of four under-expressed proteins (keratin 14, keratin 16, a protein similar to hormerin, and keratin 17) in EBC from normal subjects. Keratin 14 is expressed in the basal cells, whereas keratin 17 is expressed in the basal and some suprabasal cells. Keratin 16 is thought not to be present in normal bronchial epithelium. Wetzels et al. [24] also found that antibodies to basal cell keratins 14 and 17 displayed positivity only in squamous cell carcinomas, although no correlation with the degree of differentiation could be observed. Keratin 16 appeared to be a marker of the squamous phenotype rather than of hyperproliferation. These findings are likely to be useful in identifying lung cancer subtypes. As our results show, keratin 14 was only expressed in squamous cell carcinoma. We suggest that keratin 14, 16 and 17 are derived from the lung tissue and not from ambient air. There may also be an association between keratin 14 and squamous cell carcinoma. Moreover, these proteins may be downregulated in other lung cancer subtypes. The functions of these proteins in the carcinogenesis of lung cancer have yet to be studied and more samples are required to confirm these results, but may provide useful information if confirmed to be associated with lung cancer.
We detected the presence of growth hormone regulated TBC protein 1, which has not been found in previous studies. Growth hormone regulated TBC protein 1 is a 258 amino acid protein that contains the TBC signature motif of GTPase activator proteins of Rab-like small GTPases. As it was detected only in one lung cancer patient, further investigation on a larger scale study is required to confirm this observation. It also highlights the heterogeneous nature of the disease which has been shown in recent genomic studies. It is highly likely that an array of markers would need to be used.
Although all subjects were recruited prior to treatment (e.g. radiation or chemotherapy), most of the lung cancer patients had advanced disease at presentation (i.e. Stage III-IV). Thus, our results may not be directly applicable to the use of EBC analysis as a screening tool for early detection of lung cancer in high risk populations, and further research is necessary with subgroup analysis of earlier stage disease. Future studies could include increasing the sample size to allow for analysis of lung cancer histopathology sub-types and the effect of disease stage.
In conclusion, this study has shown significant differences in protein profiling between lung cancer subjects and healthy control groups. If our data are confirmed in a larger number of patients, the proteomic analysis of EBC could have the potential to provide clinicians with a panel of biomarkers as a simple, non-invasive screening tool for the early detection of lung carcinomas.
5. Conflict of Interest
There is no conflict of interest with regard to this manuscript.
6. Acknowledgements
Mass spectrometric results were obtained at the Bioanalytical Mass Spectrometry Facility within the Analytical Centre of the University of New South Wales. This work was undertaken using infrastructure provided by NSW Government co-investment in the National Collaborative Research Infrastructure Scheme (NCRIS). Subsidised access to this facility is gratefully acknowledged. The authors gratefully acknowledge Beth Ivimey for assisting in patient recruitment.
REFERENCES
- A. Jemal, R. Siegel, E. Ward, Y. Hao, J. Xu, T. Murray and M. J. Thun, “Cancer Statistics,” CA Cancer Journal for Clinicians, Vol. 58, No. 2, 2008, pp. 71-96. doi:10.3322/CA.2007.0010
- A. Jemal, R. Siegel, E. Ward, T. Murray, J. Xu and M. J. Thun, “Cancer Statistics,” CA Cancer Journal for Clinicians, Vol. 57, No. 1, 2007, pp. 43-66. doi:10.3322/canjclin.57.1.43
- M. J. Hayat, N. Howlader, M. E. Reichman and B. K. Edwards, “Cancer Statistics, Trends, and Multiple Primary Cancer Analyses from the Surveillance, Epidemiology, and End Results (SEER) Program,” Oncologist, Vol. 12, No. 1, 2007, pp. 20-37. doi:10.1634/theoncologist.12-1-20
- R. D. Petty, M. C. Nicolson, K. M. Kerr, E. Collie-Duguid and G. I. Murray, “Gene Expression Profiling in Non-small Cell Lung Cancer: From Molecular Mechanisms to Clinical Application,” Clinical Cancer Research, Vol. 10, No. 10, 2004, pp. 3237-3248. doi:10.1158/1078-0432.CCR-03-0503
- M. Reck, F. Crin and Lucio, “Advances in Anti-VEGF and Anti-EGFR Therapy for Advanced Non-small Cell Lung Cancer,” Lung Cancer, Vol. 3, No. 1, 2009, pp. 1-9. doi:10.1016/j.lungcan.2008.05.015
- L. L. Humphrey, S. Teutsch, M. Johnson and U. S. P. S. T. Force, “Lung Cancer Screening with Sputum Cytologic Examination, Chest Radiography and Computed Tomography: An update for the U.S. Preventive Services Task Force,” Annals of Internal Medicine, Vol. 140, No. 9, 2004, pp. 740-753.
- M. F. Reed, M. Molloy, E. L. Dalton and J. A. Howington, “Survival after Resection for Lung Cancer is the Outcome that Matters,” American Journal of Surgery, Vol. 188, No. 5, 2004, pp. 598-602. doi:10.1016/j.amjsurg.2004.07.037
- D. H. Conrad, J. Goyette and P. S. Thomas, “Proteomics as a Method for Early Detection of Cancer: A Review of Proteomics, Exhaled Breath Condensate, and Lung Cancer Screening,” Journal of General Internal Medicine, Vol. 23, Suppl. 1, 2008, pp. 78-84. doi:10.1007/s11606-007-0411-1
- J. Liu and P. S. Thomas, “Exhaled Breath Condensate as a Method of Sampling Airway Nitric Oxide and Other Markers of Inflammation,” Medical Science Monitor, Vol. 11, No. 8, 2005, pp. MT53-62.
- T. M. Dwyer, “Sampling Airway Surface Liquid: Non-volatiles in the Exhaled Breath Condensate,” Lung, Vol. 182, No. 4, 2004, pp. 241-250. doi:10.1007/s00408-004-2506-3
- K. W. Garey, M. M. Neuhauser, R. A. Robbins, L. H. Danziger and I. Rubinstein, “Markers of Inflammation in Exhaled Breath Condensate of Young Healthy Smokers,” Chest, Vol. 125, No. 1, 2004, pp. 22-60. doi:10.1378/chest.125.1.22
- E. Dalaveris, T. Kerenidi, A. Katsabeki-Katsafli, T. Kiropoulos, K. Tanou, K. I. Gourgoulianis and K. Kostikas, “VEGF, TNF-Alpha and 8-Isoprostane Levels in Exhaled Breath Condensate and Serum of Patients with Lung Cancer,” Lung Cancer, Vol. 64, No. 2, 2009, pp. 219-225. doi:10.1016/j.lungcan.2008.08.015
- G. E. Carpagnano, A. Spanevello, C. Curci, F. Salerno, G. P. Palladino, O. Resta, G. Di Gioia, F. Carpagnano and M. P. Foschino Barbaro, “IL-2, TNF-alpha, and Leptin: Local Versus Systemic Concentrations in NSCLC Patients,” Oncology Research, Vol. 16, No. 8, 2007, pp. 375- 381.
- J. Liu and P. S. Thomas, “Relationship between Exhaled Breath Condensate Volume and Measurements of Lung Volumes,” Respiration, Vol. 74, No. 2, 2007, pp. 142-145. doi:10.1159/000094238
- M. J. Raftery, “Enrichment by Organomercurial Agarose and Identification of Cys-Containing Peptides from Yeast Cell lysates,” Analytical Chemistry, Vol. 80, No. 9, 2008, pp. 3334-3341. doi:10.1021/ac702539q
- C. L. Gatlin, G. R. Kleemann, L. G. Hays, A. J. Link and J. R. Yates, “Protein Identification at the Low Femtomole Level from Silver-Stained Gels Using a New Fritless Electrospray Interface for Liquid Chromatography-MiCrospray and Nanospray Mass Spectrometry,” Analytical Biochemistry, Vol. 263, No. 1, 1998, pp. 93- 101. doi:10.1006/abio.1998.2809
- R. M. Effros, J. Biller, B. Foss, K. Hoagland, M. B. Dunning, D. Castillo, M. Bosbous, F. Sun and R. Shaker, “A Simple Method for Estimating Respiratory Solute Dilution in Exhaled Breath Condensates,” American Journal of Respiratory & Critical Care Medicine, Vol. 168, No. 12, 2003, pp. 1500-1505. doi:10.1164/rccm.200307-920OC
- I. Horvath, Z. Lazar, N. Gyulai, M. Kollai and G. Losonczy, “Exhaled Biomarkers in Lung Cancer,” European Respiratory Journal, Vol. 34, No. 1, 2009, pp. 261-275. doi:10.1183/09031936.00142508
- J. Peng, J. E. Elias, C. C. Thoreen, L. J. Licklider and S. P. Gygi, “Evaluation of Multidimensional Chromatography Coupled with Tandem Mass Spectrometry (LC/LCMS/MS) for Large-Scale Protein Analysis: The Yeast Proteome,” Journal of Proteome Research, Vol.2, No.1, 2003, pp. 43-50. doi:10.1021/pr025556v
- A. S. Jackson, A. Sandrini, C. Campbell, S. Chow, P. S. Thomas and D. H. Yates, “Comparison of Biomarkers in Exhaled Breath Condensate and Bronchoalveolar Lavage,” American Journal of Respiratory Critical Care Medicine, Vol. 175, No. 3, 2007, pp. 222-227. doi:10.1164/rccm.200601-107OC
- E. Gianazza, L. Allegra, E. Bucchioni, I. Eberini, L. Puglisi, F. Blasi, C. Terzano, R. Wait and C. R. Sirtori, “Increased Keratin Content Detected by Proteomic Analysis of Exhaled Breath Condensate from Healthy Persons Who Smoke,” American Journal of Medicine, Vol. 117, No. 1, 2004, pp. 51-54. doi:10.1016/j.amjmed.2004.01.022
- C. Gessner, H. Dihazi, S. Brettschneider, S. Hammerschmidt, H. Kuhn, K. Eschrich, T. Keller, L. Engelmann, U. Sack and H. Wirtz, “Presence of Cytokeratins in Exhaled Breath Condensate of Mechanical Ventilated Patients,” Respiratory Medicine, Vol. 102, No. 2, 2008, pp. 299-306. doi:10.1016/j.rmed.2007.08.012
- H. J. Hoffmann, L. M. Tabaksblat, J. J. Enghild and R. Dahl, “Human Skin Keratins are the Major Proteins in Exhaled Breath Condensate,” European Respiratory Journal, Vol. 31, No. 2, 2008, pp. 380-384. doi:10.1183/09031936.00059707
- R. H. Wetzels, H. E. Schaafsma, I. M. Leigh, E. B. Lane, S. M. Troyanovsky, S. S. Wagenaar, G. P. Vooijs and F. C. Ramaekers, “Laminin and Type VII Collagen Distribution in Different Types of Human Lung Carcinoma: Correlation with Expression of Keratins 14, 16, 17 and 18,” Histopathology, Vol. 20, No. 4, 1992, pp. 295-303. doi:10.1111/j.1365-2559.1992.tb00986.x

