Advances in Bioscience and Biotechnology
Vol.4 No.5A(2013), Article ID:31976,16 pages DOI:10.4236/abb.2013.45A001
Sequence and structure of naturally-occurring tRNA transcripts and site-directed variants are significant barriers to forming oligomers beyond dimers
![]()
Department of Biochemistry, University of Otago, Dunedin, New Zealand
Email: *harold.bernhardt@otago.ac.nz
Copyright © 2013 Harold S. Bernhardt, Warren P. Tate. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 1 April 2013; revised 5 May 2013; accepted 20 May 2013
Keywords: Transfer RNA; TRNA; Dimerization; Oligomerization; Kissing-Loop Interaction
ABSTRACT
Dimers of tRNAs can form through quasi self-complementary anticodon-anticodon interactions, for example at neutral pH in yeast tRNAAsp(GUC) and at pH 4.5 in Escherichia coli tRNAGly(GCC) through a partially protonated interaction. The requirements for tRNA oligomerization, and the factors that prevented higher orders of structures forming were examined with unmodified wild-type and variant E. coli tRNAsGly(GCC). Non-denaturing agarose gel electrophoresis was used as a rapid screening method. A number of tRNAGly(GCC) variants with nucleotide substitutions in the loop regions formed dimers, but surprisingly there was no evidence that distinct higher oligomers formed in any of the variants tested. The dimer interfaces of two of the variants were delineated by competitive inhibition with complementary DNA oligonucleotides. Components of an oligomerization facilitating buffer, containing monovalent, divalent and multivalent cations (magnesium and sodium ions and spermine), were tested separately and in combination, to optimize oligomerization and its detection using agarose gel electrophoresis. A rationale for the requirement for magnesium for dimerization is suggested from its role in RNA looploop interactions. Sequence specific variant tRNAs that can rapidly form heterodimers with damaging infectious RNA are potential therapeutic agents against viral mechanisms by acting as base pairing inhibitors.
1. INTRODUCTION
Dimerization of tRNAs occurs between naturally-occurring post-transcriptionally modified single tRNAs, between tRNAs in heterogenous mixtures of tRNA species [1], and also between unmodified tRNAs synthesized by in vitro transcription [2,3]. A number of post-transcriptionally modified tRNAs form dimers in their crystal structures, either through an anticodon-anticodon interaction in the cases of yeast tRNAAsp(GUC) [4-6] and E. coli Cys-tRNACys [7], or an interaction between the anticodon and the 3’ CCA terminus in the case of tRNALys(s2UUU) [8]. Yeast tRNAAsp(GUC) associates through its quasi selfcomplementary GUC anticodon to form dimers both in the crystal [4,5] and in solution [9], although in solution the interaction is quite weak, occurring only at low temperature and neutral pH [9]. Escherichia coli tRNAGly(GCC) also contains a quasi self-complementary anticodon—in this case GCC—but in contrast only forms dimers in solution at pH 4-5 [10]. This dimerization can be explained by partial protonation of the GCC anticodon of tRNAGly(GCC) at acidic pH, with the two central cytosines sharing a single proton in a protonated base pair. C-C(+) interactions are well documented in DNA dinucleotide repeats [11] and trinucleotide repeats [12] and in turnip yellow mosaic virus RNA [13]. Although the structure of tRNAGly(GCC) from yeast has been elucidated [14], crystallization required a high concentration of organic solvent (50% dioxane-water (v/v)), and an acidic pH of 6.1. The crystal structure was unusual in that the acceptor stem was splayed open, but there was no evidence of dimer formation, as might have been predicted at this higher pH. It has been suggested by Dirheimer and colleagues [15] that the crystals in fact contained a mixture of tRNA species.
Unmodified tRNAs can be produced by in vitro transcription and have similar, if somewhat looser, 3D structures compared to their respective post-transcriptionally modified native versions [16-20]. Dimerization of a variant of human mitochondrial tRNALeu(UAA) (A14G) has been implicated in the morbidity of MIDD (maternally-inherited diabetes and deafness) and MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) [21]. The A14G mutation creates a self-complementary GGGCCC sequence in the D arm of the tRNA, which, in experiments with the unmodified in vitro transcribed tRNA, Kelley and colleagues showed was responsible for dimer formation [21,22]. The nucleotides of the A14G mutant tRNA at the GGGCCC dimer interface are unmodified in vivo, and modification of the nearest modified nucleotide is markedly reduced. Wittenhagen and Kelley argue that the unmodified D stem of their unmodified transcripts is analogous to the population of the human mitochondrial tRNALeu(UAA) in cells [21].
Kholod divides tRNA dimers into two categories: “true” dimers and dimer complexes [1]. According to this definition, our work focuses on the second category: labile assemblies formed through loop-loop or “kissing” base pair interactions that are sensitive to changes in solution conditions, such as pH and Mg2+ concentration. Most tRNA dimer complexes form at neutral pH and dimerization can be initiated if there is a Mg2+ binding site at the interface. An example is the dimerization of the A14G mutant human mitochondrial tRNALeu(UAA) that was dependent on Mg2+ (4 mM promoted 50% dimerization) [22]. Kelley and colleagues [22] suggest that this requirement for Mg2+ is similar to that for the HIV-1 DIS dimerization where a Mg2+-binding site may also exist at the dimer interface [23]. Temperature, and the concentrations of monovalent cations and multivalent cations such as spermine and spermidine are other important variables for dimer formation [1]. The A14G mutant human mitochondrial tRNALeu(UAA) dimer has a higher rate of formation at 37˚C than 25˚C, perhaps due to destabilization of the D stem—and consequent exposure of the selfcomplementary hexanucleotide sequence—at the higher temperature [21]. The requirement for monovalent and multivalent cations for tRNA dimer formation is more system specific, with 1.6 M 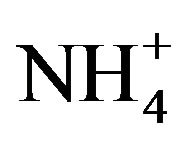 plus 3mM spermine (but without Mg2+) increasing the stability of the yeast tRNAAsp(GUC) dimer [9], and Li+ or Na+ or K+ (0.5 M) producing a low level of dimerization of A14G mutant human mitochondrial tRNALeu(UAA) [22].
plus 3mM spermine (but without Mg2+) increasing the stability of the yeast tRNAAsp(GUC) dimer [9], and Li+ or Na+ or K+ (0.5 M) producing a low level of dimerization of A14G mutant human mitochondrial tRNALeu(UAA) [22].
Of the four single-stranded regions of tRNA, the anticodon loop is the most common interface for tRNA dimerization. This is not surprising, given the anticodon’s primary role in protein synthesis as an exposed sequence that naturally forms base pair interactions with mRNA on the ribosome. In Gram positive bacteria the anticodons of some tRNAs interact with the 5’ UTR of a number of regulated amino acid-related genes, the transcription of which is regulated by the relative amounts of the specific aminoacylated vs deacylated tRNA [24,25]. The single-stranded CCA sequence of the 3’-end of tRNA also forms intermolecular interactions in vivo, with 23S rRNA [26,27], RNase P RNA [28], and with the 5’ UTR of regulated amino acid-related genes in Grampositive bacteria [24,25,29,30]. The D and T loop of tRNA are not so exposed as they are primarily involved in intramolecular base pair interactions that cement the corner of the L-shaped tertiary structure of the molecule. The two main D loop-T loop interaction—G18 (D loop)-Y55 (T loop) and G19 (D loop)-C56 (T loop)—are trans-Hoogsteen base pairs [31]. Even with these pairs, however, it has been demonstrated that the tight coupling between the two loops may have some flexibility. In the two crystal structures obtained of the tRNAAsp dimer, the G19 (D loop)-C56 (T loop) base pair is either very weak or absent [6]. Moras and colleagues hypothesize that this may be due to a transfer of mobility from the anticodon loop to the D/T loop corner upon anticodon-anticodon binding [6], suggesting a degree of cooperativity in tRNA intermolecular interactions. And in a paper entitled “tRNA prefers to kiss” [32], Scarabino and colleagues evolved RNA aptamers that would bind a post-transcriptionally unmodified in vitro transcript of yeast tRNAPhe, one of which contained two hexanucleotide sequence blocks complementary to all seven D loop nucleotides and all eight T loop nucleotides, respectively. This aptamer bound even more strongly to a tRNAPhe in which the D loop/T loop tertiary interaction had been disrupted by U8A and G18C mutations [32]. The above examples show that tRNA is able to form multiple intermolecular interactions simultaneously.
How does RNA structure facilitate dimerization generally? The HIV-1 dimerization-initiation site (DIS) consists of a stem-loop structure with a self-complementary GCGCGC or GUGCAC loop sequence, and avian leukosis virus RNA [33] has the self-complementary loop sequence GGGCCC, the same as in the A14G mutant human tRNALeu(UAA) [21,22]. The dimers formed by both GCGCGC sequences have similar stability [34]. Harada and associates synthesized novel RNA hairpins with loops containing complementary sequences based on the GCGCGC sequence found in the HIV-1 subtype B DIS, and found these oligomerized into multimeric products including linear and circular arrangements [35-37]. The formation of multimers was higher at lower temperatures (4˚C c.f. 23˚C) with 0.1 mM Mg2+ (the buffer also contained 10 mM sodium phosphate and 50 mM NaCl at pH 7.0) [36]. Jaeger and colleagues used hybrid glutamine/serine tRNAs containing a long variable arm to form RNA squares, ladders, triads, 3D prisms and polyhedrons [38,39]. They doubled the length of the anticodon and acceptor stems, and circularly permuted the sequence in order to form an additional loop at the end of the acceptor stem between the tRNA 3’ and 5’ ends, inserting complementary (but not self-complementary) sequences of six nucleotides into the anticodon and (new) acceptor stem loops to allow formation of intermolecular base pair interactions. Computer modeling was used to adjust the length and orientation of the stem/loops of their altered tRNA building blocks to optimize base pair interactions. Oligomerization of these hyper-modified variants was dependent upon Mg2+.
For our study, we synthesized a range of E. coli tRNAGly(GCC) site-directed sequence variants to deduce the minimum requirement(s) for oligomerization in the template tRNAs without addition of extra structural elements. E. coli tRNAGly(GCC) was chosen to model the effects as it possessed a number of complementary sequences in its loops and single-stranded regions. We investigated whether disrupting the intramolecular D loopT loop base pairs would facilitate the complementary sequences base pairing with other molecules.
2. MATERIALS AND METHODS
2.1. tRNA Sequences and Secondary Structure Prediction
The E. coli tRNAGly(GCC) gene sequence was obtained from the ncbi structural RNA database at http://www. ncbi.nlm.nih.gov. Analysis of unmodified tRNA secondary structures was carried out online using the RNAfold website http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi [40- 42]. Folding temperature used was 4˚C. Structures shown are minimum free energy (MFE) secondary structures. Base pair probabilities are shown by a colour spectrum, with red most probable and blue least probable.
2.2. Synthesis of tRNA Templates
Recombinant pUC18 vector containing human mitochondrial WT tRNALeu(UAA) was the gift of Dr Lisa Wittenhagen, Boston College, USA. Amplification of the E. coli tRNAGly(GCC) gene from genomic E. coli DNA was carried out using internal primers complementary to the 5’ and 3’ ends of the tRNA gene. The forward primer contained the 17 nucleotide T7 RNA polymerase promoter sequence to allow for subsequent in vitro transcription. One-step site-directed mutagenesis PCR using the Expand High Fidelity PCR System (Roche Diagnostics GmbH) was used to introduce site-directed mutations at desired positions. All DNA primers used for PCR reactions were purchased from Invitrogen™ Life Technologies, NZ. PCR products were purified using the QIAquickÒ PCR Purification Kit (Qiagen) and, following sequential restriction endonuclease digestion with Bam HI and Pst I (Roche Diagnostics GmbH), used for cloning into the pUC18 vector. Ligations were carried out in a 20 µl reaction volume containing 1 U of T4 DNA Ligase (Roche Diagnostics GmbH) and 20 ng plasmid DNA at 14˚C - 18˚C for 11 - 24 h. Molar ratio of vector to insert was 1:8. Transformations were done using heat-shocked competent E. coli strain DH5a cells (Invitrogen Life Technologies, Germany). Plasmids were sequenced to confirm the presence of the correct variant tRNA sequence. PCR products of transformed plasmids were digested with either Mva I or Hae II restriction endonucleases (Roche Diagnostics GmbH or New England Biolabs) to linearize the plasmid and generate the correct 5’ terminus of the DNA template. Hae II was used for the variants in which the presence of a selfcomplementary GGGCCC sequence in the anticodon loop would have resulted in cleavage at an internal CCA sequence and a half-length gene product when cut by Mva I. To remove the 3’ overhang left by digestion with Hae II, DNA templates were incubated with Klenow enzyme (DNA polymerase large fragment) (Roche Diagnostics GmbH) in a reaction mix including T7 RNA polymerase buffer but excluding rNTPs and T7 RNA polymerase, prior to in vitro transcription.
2.3. In Vitro RNA Synthesis
In vitro transcription was carried out using the Ribo MAXÔ Large Scale RNA Production System-T7 (Promega) using reaction volumes of 30 - 150 µl. The supplied protocol was modified to give an increased concentration of T7 RNA polymerase, with a corresponding reduction in the concentration of rNTPs and template DNA. Following the protocol of Kelley and colleagues [22], a second aliquot of T7 RNA polymerase was added to the incubation after 3 h, with incubation continued overnight (13 - 21 h) at 37˚C. T7 RNA polymerase was from the RiboMAXÔ kit (Promega) or a laboratory preparation expressed from a cloned gene; the latter was used with RNasin® Plus RNase Inhibitor (from RiboMAXÔ kit) or RNaseOUTÔ Recombinant Ribonuclease Inhibitor (Invitrogen Life Technologies, USA). T7 RNA polymerase buffer (5x) was from the RiboMAXÔ kit (Promega) or prepared in the laboratory by HSB. The in vitro transcription reaction was incubated at 37˚C for 16 - 24 h to produce post-transcriptionally unmodified tRNA transcripts. As recommended in the Promega protocol, following in vitro transcription the DNA template was removed by a 15 - 30 min incubation at 37˚C with RQ1 RNase-free DNase (from RiboMAXÔ kit) at >1 U RQ1/µg template DNA. The transcripts were purified from the in vitro transcription mix using phenol-chloroform extraction. TRIZOL Reagent (Invitrogen Life Technologies, USA) (200 µl) and 40µl chloroform were used to extract 30 - 150 µl of an in vitro transcription mixture. Following centrifugation at 12,000 x g, the top aqueous layer was removed and loaded in 50 µl aliquots onto pre-spun illustraÔ MicrospinÔ G-25 size-exclusion columns (GE Healthcare).
2.4. tRNA Oligomerization Protocol at pH 7.5
The oligomerization protocol was adapted from Wittenhagen and Kelley [21] and involved adding the tRNA transcripts (100 - 400 ng/µl as measured using an ND- 1000 spectrophotometer (NanoDrop, USA)) in sterilized MilliQ water to a thin-walled 0.5 ml PCR tube containing 5× incubation buffer (1 × 10 mM Tris-HCl pH 7.5), with mixing. The tube was then heated to 95˚C for 5 min using a PTC-200 Peltier Thermal Cycler (MJ Research, USA) to denature the tRNA secondary structure and remove potential aberrant structures, and then immediately cooled on wet ice for 2 - 5 min to renature the tRNA. To induce oligomerization, tRNA transcripts were then transferred to a thin-walled 0.5 ml PCR tube containing either 5× oligomerization buffer to make the final solution either 10 mM Mg2+, 1 mM spermine and 100 mM NaCl (MSN buffer), or 10 mM Mg2+ and 100 mM NaCl (MN buffer). After mixing, incubation was for the times noted at 4˚C. Concentrations of tRNA variants were as indicated for the individual experiments.
2.5. Native Agarose Gel Electrophoresis
Following oligomerization experiments, tRNA transcripts were separated by non-denaturing agarose gel electrophoresis on a 2% (w/v) agarose gel in TBM (89 mM Tris, 89 mM boric acid, 0.1 mM MgCl2) or 0.5 TBE buffer (44.5 mM Tris, 44.5 mM boric acid, 1.3 mM EDTA) containing 0.5 µg/ml ethidium bromide, in running buffer containing TBM or 0.5 TBE with 0.05 µg/ml ethidium bromide added at the cathode (+ ve) end with mixing. Following oligomerization experiments, tRNA samples (2.6 - 7.5 µM tRNA/lane) were mixed with non-denaturing agarose gel loading dye (50% (w/v) sucrose, 0.1% (w/v) bromophenol blue). RNA CenturyÔ Plus Size Markers (Ambion), containing sequences of 100, 200, 300, 400, 500, 750 and 1000 nucleotides were mixed with denaturing agarose gel loading dye (50% (w/v) sucrose, 7 M urea, 0.1 mM EDTA (pH 8.0), 0.1% (w/v) bromophenol blue), heated at 95˚C for 5 min using a PTC-200 Peltier Thermal Cycler (MJ Research, USA), and loaded onto the gel immediately. The gels were run at 4˚C (40 V constant voltage or 17 mA constant current) or RT (90 V constant voltage) for 1 - 5 h, and the tRNAbound ethidium bromide detected with UV light on a Gel Doc 2000ä gel documentation system (Bio-Rad Laboratories, USA).
2.6. Oligonucleotide Inhibition Protocol
The oligonucleotide inhibition protocol was adapted from Wittenhagen and Kelley [21]. DNA oligonucleotides with sequences complementary to various regions of the variant tRNAs were from Invitrogen™ Life Technologies, NZ. These oligonucleotides were reconstituted at 1 nmol/µl and added to the tRNA transcript to give a molar ratio of oligonucleotide to tRNA of 20:1. The tRNA/oligonucleotide mixtures were used in the oligomerization protocol as described above. Incubation times were as noted.
3. RESULTS
The aim of our study was to investigate the potential for naturally occurring tRNA transcripts to form higher order structures. In contrast to Jaeger and colleagues, who used drastically altered tRNA structures in their work [38], [39], we were interested in determining whether the standard tRNA scaffold itself would be suitable as a building block. Like H2O, tRNA has tetrahedral symmetry [43]. To take advantage of this shape to form longrange hydrogen bonded super-molecular complexes and to utilize all four single-stranded regions of tRNA for intermolecular interactions, the D loop-T loop base pairing interaction was disrupted by site-directed mutagenesis, so that variants of the tRNA scaffold contained mutations in the T loop. The D and T loops were then free to interact with other molecules and provide interfaces for intermolecular interactions. Variants containing mutations in the D and anticodon loops, and in multiple loops, were also analyzed.
Although ultimately sophisticated analytical techniques such as atomic force microscopy (AFM) would be required for detailed analysis of any oligomeric structures formed, we used agarose gel electrophoresis as a rapid screening methodology for detecting oligomers of tRNA. Agarose gel electrophoresis has not to our knowledge been used previously to study tRNA oligomerization, but has been utilized for studies of HIV-1 genome dimerization [44]. Wittenhagen and Kelley by contrast used non-denaturing PAGE in their study of A14G mutant human mitochondrial tRNALeu(UAA) dimerization [21], however such gels tend to be fragile and not always reproducible. To promote oligomerization we evaluated an MSN buffer (10 mM Mg2+, 1 mM spermine and 100 mM NaCl) [21] or the buffer lacking spermine (MN). Initially MSN buffer was used, following the protocol of Wittenhagen and Kelley for their studies of unmodified A14G mutant human mitochondrial tRNALeu(UAA) dimerization [21]. In these studies, the dimer was found to be strongly stabilized by 1 - 3 mM spermine (L. Wittenhagen, pers. commun.). By contrast, spermine was not required for dimerization of our tRNA variants, and it tended to produce a more diffuse dimer band on the agarose gel. MN buffer was therefore used for the later oligomerization experiments.
No studies have previously been published using posttranscriptionally unmodified transcripts of E. coli tRNAGly(GCC) apart from our two preliminary communications predicating the present work [43,45]. Although unmodified tRNAs have a somewhat looser 3D structure compared with their post-transcriptionally modified versions [16-20], the expectation is that the unmodified wild-type E. coli tRNAGly(GCC) scaffold possesses a typical tRNA clover-leaf secondary structure and Lshaped tertiary structure, including a D loop-T loop base pair between G18 and U55 (U55 is post-transcriptionally modified to Ψ55 in vivo). A cloverleaf secondary structure representation of unmodified wild-type E. coli tRNAGly(GCC) is shown in Figure 1, annotated with the introduced mutations used to create the variants in this study. Although in Figures 1(c) and (d) the variants are shown as typical cloverleafs, this is for comparative purposes only, as the T- > anti-D and self-complementary variants are predicted by RNAfold to have different secondary structures.
3.1. Oligomerization Time-Course of Wild-Type tRNAGly(GCC) at pH 7.5
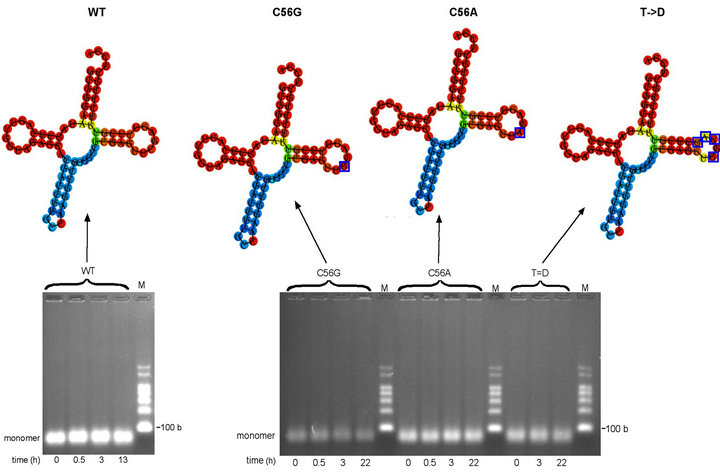
Romby and colleagues [10] showed that post-transcriptionally modified wild-type E. coli tRNAGly(GCC) does not form dimers at neutral pH. Unmodified wild-type E. coli tRNAGly(GCC) was produced in vitro and subjected to a standardized oligomerization protocol (see Materials and Methods) with aliquots incubated at pH 7.5 and 4˚C for 0, 0.5, 3 h and overnight. The results, shown in Figure 2(a), exhibited no evidence of oligomerization even after 13 h incubation. Although agarose gel electrophoresis was attempted at pH 5.0, the bands could not be visualized, as ethidium bromide does not fluoresce at acidic pH. The RNAfold secondary structure prediction for unmodified wild-type (WT) E. coli tRNAGly(GCC) is shown in Figure 2(a) (upper). This is consistent with the standard cloverleaf secondary structure (see Figure 1) except for the anticodon loop, shown as just three nucleotides, consisting only of the anticodon itself. Complementary nucleotides on the 3’ and 5’ sides of the loop are shown as being base paired. The anticodon loop of WT E. coli tRNAGly(GCC) is not post-transcriptionally modified in vivo, supporting the model of a three nucleotide anticodon loop—and seven base pair anticodon stem—as has been found to be the case for other tRNAs [46-48].
3.2. Oligomerization Time-Course of C56G C56A and T - > D Variants
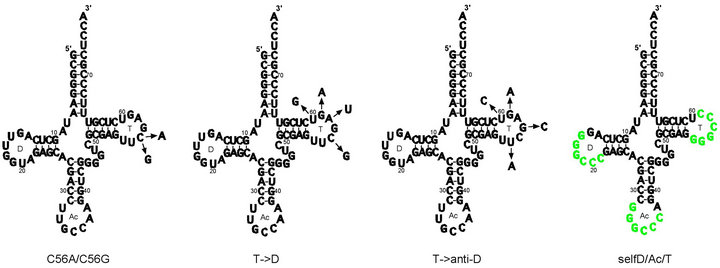
Following the lack of observed oligomerization for wildtype E. coli tRNAGly(GCC), we synthesized five variants of wild-type tRNAGly(GCC) by site-directed mutagenesis (see Materials and Methods); cloverleaf secondary structure representations of wild-type E. coli tRNAGly(GCC) showing the position of mutations in these variants are depicted in Figure 1. The C56G variant (Figure 1(a)) was designed to test whether disrupting the G19-C56 base pair would be sufficient to disrupt the D loop-T loop interaction at the corner of the molecule (as noted, along with the G18-U55 base pair in the unmodified tRNA, G19-C56 is one of two D loop—T loop tertiary base pairs that cement the elbow of tRNA’s L-shaped struc-
 (a) (b) (c) (d)
(a) (b) (c) (d)
Figure 1. C56G, C56A, T- > D, T- > anti-D variants and selfD/Ac/T variants shown in relation to wild-type E. coli tRNAGly(GCC) cloverleaf secondary structure. Mutations introduced to produce (a) C56G and C56A variants; (b) T- > D variant; (c) T- > anti-D variant; (d) Self-complementary GGGCCC sequences are shown in green, indicating positions in the various self-complementary variants (e.g. selfT, selfD, selfAc etc). NB: cloverleaf representations of T- > anti-D and selfD/Ac/T variants indicate the positions of the mutations only, as these variants do not apparently form standard cloverleaf secondary structures. D = D loop, Ac = anticodon loop, T = T loop. Wild-type sequence taken from Jühling et al. [42].
 (a) (b)
(a) (b)
Figure 2. Oligomerization time-courses of wild-type (WT) tRNAGly(GCC) and C56G, C56A and T- > D variants (a) Minimum free energy (MFE) secondary structure of WT tRNAGly(GCC) produced using RNAfold online programme shown above oligomerization time-course of WT tRNAGly(GCC) transcript in MSN oligomerization buffer at 4˚C. WT tRNAGly(GCC) transcript at 5.7 µM. (b) Minimum free energy (MFE) secondary structures of variant tRNAsGly(GCC) produced using RNAfold online programme (mutated nucleotides in T loops indicated by blue boxes) shown above oligomerization time-course study of C56G, C56A and T- > D variants in MSN oligomerization buffer at 4˚C. C56G variant transcript at 4.7 µM. C56A variant transcript at 5.7 µM. T- > D variant transcript at 5.1 µM. Predicted size of all transcripts: 76 bases. M = RNA markers. b = bases.
ture), while at the same time increasing the complementarity of the resulting T loop for the anticodon loop and CCA terminus. The C56A variant (Figure 1(a)) would have a disrupted G19-C56 interaction without increasing the complementarity of the T loop sequence. This variant would test whether disruption of G19-C56 alone would facilitate oligomerization by allowing the D loop UGG sequence including G19 to interact with complementary CCA sequences both within the anticodon loop and at the 3’-end. Interactions would involve only three base pairs, but two would be G-C. There are a number of examples of RNA oligomerization involving only two G-C base pairs, such as in yeast tRNAAsp(GUC) dimerization (where the central uridines are thought not to base pair [4,5]), the dimerization of the Moloney murine leukemia virus tetraloop [49] and in the proposed reading of codons by tRNA using only two nucleotides (two-out-of-three mechanism) [50-53]. The T- > D variant (Figure 1(b)) was designed as an alternative to make the T loop identical in sequence to six nucleotides of the D loop plus one from the adjacent D stem. As with the C56G and C56A variants, these mutations are expected to disrupt the intramolecular G19-C56 interaction but not the G18-U55 interaction, making the T loop complementary to all seven nucleotides of the anticodon loop (including a G-U base pair). The unmodified C56G, C56A and T- > D variants of tRNAGly(GCC) were put through the standard oligomerization protocol (see Materials and Methods) with aliquots left at 4˚C for 0, 0.5, 3 h and overnight. Under the conditions of this assay there was no evidence of oligomerization for any of the three variants, even following overnight incubation (Figure 2(b)). The lack of dimers or higher structures for the C56G, C56A and T- > D variants, suggested that disruption of the G19-C56 base pair is alone either insufficient to disrupt the D loop-T loop intramolecular interaction or the potentially complementary sequences are insufficient to form stable base pair interactions with other molecules.
3.3. Oligomerization Time-Course of T- > Anti-D Variant
The T- > anti-D variant was designed to make the T loop complementary to seven nucleotides of the D loop by the mutation of four nucleotides of the T loop (the mutations are shown in Figure 1(c); as indicated previously, use of the cloverleaf secondary structure is for illustrative purposes only). In the reverse situation to the C56G, C56A and T- > D variants, these mutations were expected to result in disruption of the G18-U55 base pair (in the unmodified transcript) but not the G19-C56 base pair. This variant tested whether disrupting this base pair would be successful in separating the D and T loops and make possible the potentially complementary intermolecular interactions. It was designed so that the presence of two sets of complementary sequences (in the D and T loops) in this variant would enable the formation of oligomers higher than dimers. The results of oligomerization time-course experiments were surprising: as shown in Figure 3(a), while the T- > anti-D variant did indeed form dimers, there was no evidence of distinct species of higher oligomers. Dimer formation proceeded slowly over time, first evident at 30 min (Figure 3(a), Lane 2;
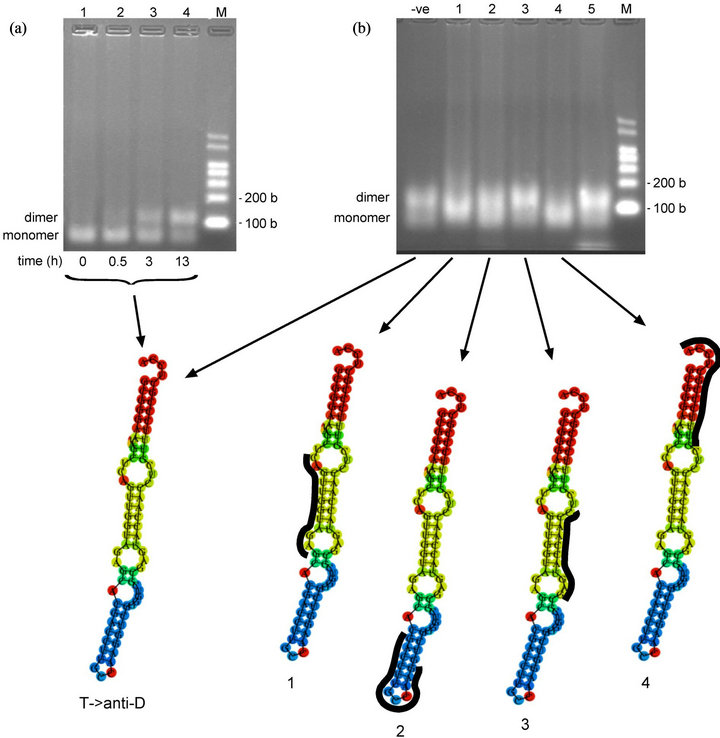
Figure 3. Oligomerization time-course and oligonucleotide-inhibition study of T- > anti-D variant (a) Oligomerization time-course of T- > anti-D variant in MSN oligomerization buffer at 4˚C. T- > anti-D transcript at 4.1 µM. Predicted size of T- > anti-D transcript: 76 bases. (lower) Minimum free energy (MFE) secondary structure produced online via the RNAfold website (b) Oligonucleotide-inhibition study of T- > anti-D variant in MSN oligomerization buffer for 10.5 h at 4˚C. T- > anti-D transcript at 3.0 µM. (lower) Minimum free energy (MFE) secondary structures showing binding positions of DNA oligonucleotides (in black) to various regions of the T- > anti-D variant. -ve control = no oligo; Lane 1, anti-D loop (WT) oligo; Lane 2, anti-Ac loop (WT) oligo; Lane 3, anti-mutated T loop (T- > anti-D variant) oligo; Lane 4, anti-CCA terminus (WT) oligo; Lane 5, anti-SCS(GGGCCC) (selfT variant) oligo (for sequences of DNA oligonucleotides, see Table 1). Predicted size of transcripts: 87 - 89 bases (76 bases tRNA + 11 - 13 bases DNA oligo). M = RNA markers. b = bases.
very faint band), but with dimer the predominant species by 13 h (Lane 4). After 4 days incubation at 4˚C there was complete conversion to the dimer, but still no evidence of higher oligomers (data not shown).
Dimerization of the T- > anti-D variant was dependent on the components of the MSN oligomerization buffer (data not shown). The RNAfold programme predicted a secondary structure for the T- > anti-D variant which departed significantly from the typical canonical clover-leaf (Figure 3(a), lower). However, RNAfold can only calculate base pair interactions within the secondary structure, and not the important tertiary interactions that affect structural stability. While it is known that the standard tRNA structure possesses up to seven tertiary interactions that greatly increase the stability of the L-shaped conformation [31], the number of stabilizing tertiary interactions (if any) in the alternative conformations as depicted by RNA-fold is unknown. Therefore, the alternative secondary structures produced by RNAfold may not always represent the lowest energy structures. Nevertheless, the lack of oligomers higher than dimers implied that the T- > anti-D variant did not possess the standard tRNA structure.
3.4. Oligonucleotide-Inhibition of T- > Anti-D Variant Oligomerization
To determine the T- > anti-D variant dimer interface, we used the protocol of Wittenhagen and Kelley [21] which involved measuring oligomerization in the presence of 11-13-mer oligonucleotides complementary to different regions of the tRNA (Figure 3(b)) (for sequences of DNA oligonucleotides, see Table 1). The result supported an altered secondary structure for this variant, with the oligonucleotide complementary to the mutated T loop sequence producing no apparent reduction in the amount of dimer, suggesting this sequence is not available for oligomerization (Figure 3(b), Lane 3). Rather, the D loop region (Lane 1) and 3’-end sequence (Lane 4) appear to be the interacting sequences responsible for oligomerization; oligonucleotides complementary to these two regions reduced dimer formation. This suggests that the D loop UGG and 3’-end CCA sequences may constitute the dimer interface. Three base pairs might not form a particularly strong interaction; however, dimer formation could involve two such interactions per dimer, meaning the base pairing would involve six nucleotides including four G-C base pairs, potentially quite a strong interacttion. Since the RNAfold programme predicted a non-cloverleaf structure for the T- > anti-D variant, a different approach was required for constructing an oligomerizing tRNA with a standard tetrahedral structure.
3.5. Oligomerization Time-Course of SelfT Variant
The selfT variant was designed to incorporate a sequence in the T loop that has a proven record of dimerization in a variety of other systems (the position of the self-complementary sequences (SCS) in the self complementary variants is shown in Figure 1(d); use of the cloverleaf secondary structure is for illustrative purposes only as with the T- > anti-D variant). The self-complementary GGGCCC hexanucleotide sequence is the dimer interface of the unmodified A14G mutant human mitochondrial tRNALeu(UAA) transcript [21,22] as well as being responsible for the dimerization of avian leukosis virus RNA [36]. We anticipated that the introduction of this sequence into the T loop should disrupt the two D loop-T loop intramolecular base pairs (G18-U55 and G19-C56 in the unmodified tRNA) that maintain the molecule’s L-shaped tertiary structure. In an initial experiment the selfT variant formed a clear dimer band almost instantly following addition of MSN in an almost complete conversion of monomer to dimer (data not shown). Because dimerization occurred so quickly, we tested whether dimerization was dependent on the ions in the MSN oligomerization buffer (Figure 4(a)). This was found to be the case, with no sign of oligomerization up to 17 hours following renaturation in the absence of MSN (Figure 4(a), Lane 4), despite oligomerization occurring extremely quickly following addition of MSN, as before (time = 0, sampling immediately after addition), with ~50% of the transcript converted to dimer (Lane 5). These results demonstrate that the GGGCCC sequence is sufficient to enable dimerization in the context of the tRNA T loop. As predicted, the presence of a single SCS limited oligomerization to formation of the dimer. RNAfold predicts the selfT SCS is incorporated into the T stem (Figure 4(a), lower) and therefore potentially less available to form intermolecular interactions.
3.6. Oligonucleotide-Inhibition of T- > Anti-D Variant Oligomerization
To confirm that the introduced GGGCCC hexanucleotide SCS is responsible for dimerization, we used the oligonucleotide inhibition protocol [21] as previously (for sequences of DNA oligonucleotides, see Table 1). Results shown in Figure 4(b) confirmed that the introduced SCS was responsible for dimerization, as the DNA oligonucleotide complementary to the region containing this sequence caused a marked decrease in dimer formation (Figure 4(b), Lane 3). Supporting this conclusion surprisingly was the fact that the anti-upstream SCS +1 oligonucleotide, which overlaps the GGGCCC sequence by just a single nucleotide on the 5’-side, also caused a decrease in the amount of dimer (Lane 6). However, the anti-downstream SCS +1 oligonucleotide, which overlaps the GGGCCC sequence by a single nucleotide on the 3’-side, failed to affect dimer formation (Lane 7).
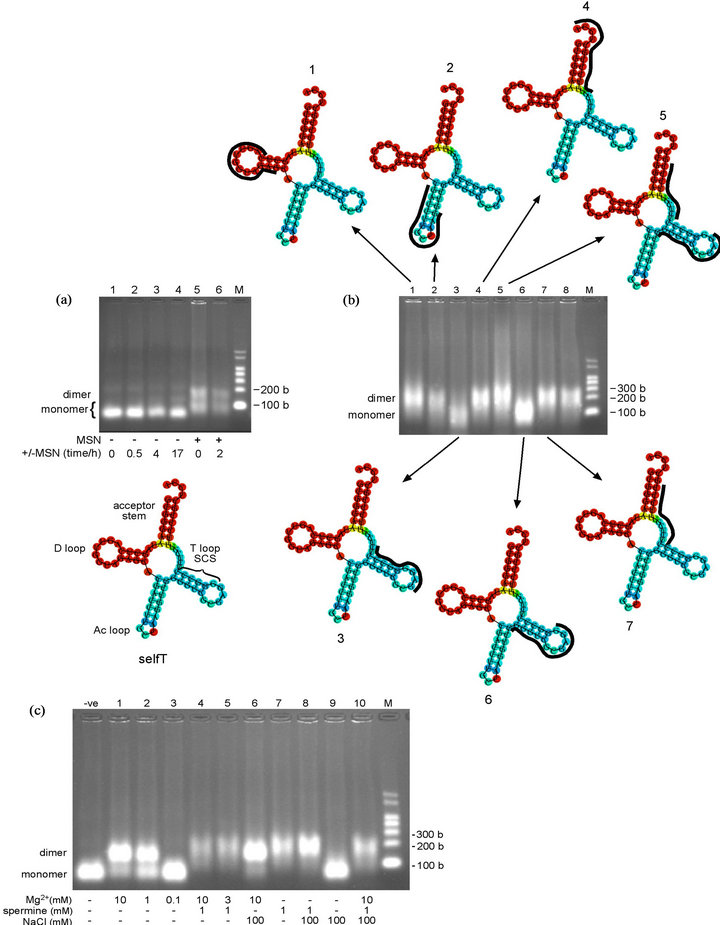
Figure 4. Oligomerization time-course and oligonucleotide-inhibition study of selfT variant, and analysis of oligomerization buffer components. (a) (upper) Oligomerization time-course of selfT variant in MSN oligomerization buffer. Lanes 1-4, heated at 95˚C for 5 min (denatured), cooled immediately on wet ice and incubated at 4˚C in the absence of MSN oligomerization buffer for the times noted; Lane 5, immediately following addition of MSN. SelfT transcript at 7.5 µM. Predicted size of selfT transcript: 76 bases. NB: The apparent molecular weight of selfT monomer increases in the presence of MSN (Lanes 5, 6 vs 1-4) (lower) Minimum free energy (MFE) secondary structure of selfT variant showing predicted location of self-complementary sequence (SCS). D = D loop, Ac = anticodon loop, T = T loop (b) (centre) Oligonucleotide-inhibition study of selfT variant in MSN oligomerization buffer for 10.5 h at 4˚C. Lane 1, anti-D loop (wild-type); Lane 2, anti-Ac loop (wild-type); Lane 3, anti-SCS (GGGCCC) (selfT variant); Lane 4, anti-CCA terminus (wild-type); Lane 5, anti-upstream SCS and anti-downstream SCS; Lane 6, anti-upstream SCS +1 (selfT); Lane 7, anti-downstream SCS +1 (selfT); Lane 8, anti-mutated T loop (T- > anti-D variant) (for sequences of DNA oligonucleotides, see Table 1). SelfT transcript at 4.6 µM. Predicted size of transcripts: 87 - 89 bases (76 bases tRNA + 11-13 bases DNA oligonucleotide) (upper/lower) Minimum free energy (MFE) secondary structure of selfT variant showing binding position of DNA oligonucleotides (in black) that do (structures 3, 6) and do not (structures 1, 2, 4, 5, 7) inhibit dimerization (c) Oligomerization of selfT variant in the presence of different concentrations and/or combinations of components of the MSN oligomerizing buffer. The selfT transcript was incubated for 14 h at 4˚C in the presence of the different combinations of agents causing oligomerization. SelfT transcript at 5.4 µM. -ve control = Tris buffer. Predicted size of transcript: 76 bases. NB: The apparent molecular weight of selfT dimer increases in the presence of spermine (Lanes 4, 5, 7, 8, 10 vs Lanes 1, 2, 6). M = RNA markers. b = bases.
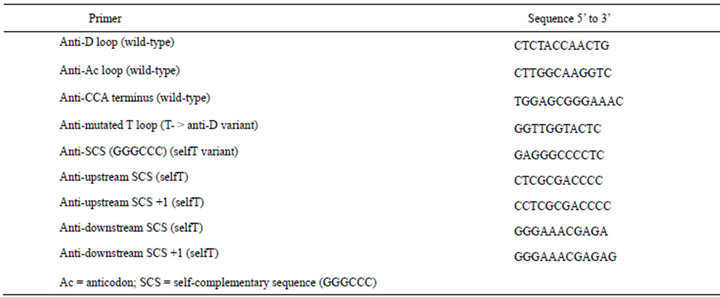
Table 1. Sequences of oligonucleotide-inhibition primers.
The combination of two oligonucleotides that abut the SCS on the 5’ and 3’ sides (anti-upstream SCS and antidownstrean SCS) supported a high level of oligomerization (Lane 5), in agreement with the findings of Wittenhagen and Kelley in similar experiments on the human mitochondrial tRNALeu(UAA) [21]. As noted above, RNAfold predicts the SCS is incorporated into the T stem (Figure 4(a), lower) and therefore is perhaps less available to form intermolecular interactions. The results of the nucleotide-inhibition study however strongly implicate the SCS in dimerization, suggesting either that the T stem is able to undergo conformational rearrangement to expose the self-complementary sequence, or the RNAfold prediction is inaccurate in this instance and the GGGCCC sequence forms part of the singlestranded T loop. As previously discussed, Kelley and colleagues [22] found a higher rate of oligomerization for A14G mutant human mitochondrial tRNALeu(UAA) at 37˚C than at 25˚C, possibly due to a requirement for denaturation of the D stem to expose the self-complementary interface. In contrast, incubation at 4˚C was used for all our experiments (including for the selfT variant), as incubation at 37˚C produced poorly differentiated bands on the agarose gel (data not shown).
3.7. Analysis of Components of MSN Oligomerization Buffer
As the selfT variant oligomerizes extremely quickly and quantitatively in the presence of MSN buffer (10 mM Mg2+, 1 mM spermine and 100 mM NaCl), we used this variant to test the individual components of MSN buffer at a range of concentrations and in different combinations, to determine their relative importance for oligomerization (Figure 4(c)). Both 1 mM Mg2+ (Figure 4(c), Lane 2) and 1 mM spermine (Lane 7) by themselves induce dimerization of the selfT variant. By comparison, NaCl alone does not (at least at 100 mM—Lane 9). However, the presence of 100 mM NaCl in combination with 1 mM spermine produces a more intense band (Lane 8 vs Lane 7). The effect of Mg2+ on oligomerization is concentration-dependent: 1 mM Mg2+ gives a lower proportion of dimer than 10 mM (Lane 2 vs Lane 1), and 0.1 mM Mg2+ produces no visible oligomerization (Lane 3). The loss of intensity of bands in the presence of spermine may be due to spermine competing with ethidium bromide (the visualizing agent used in the agarose gels) for binding to tRNA [54,55]). Oligomerization in the presence of spermine results in an increase in the molecular weight or size of the dimer (due to the binding of spermine), as can be seen from the slower running dimer band in Figure 4(c), Lanes 4, 5, 7, 8 and 10 compared with Lanes 1, 2 and 6. Following this result, we decided to omit spermine from the oligomerizing buffer and used only 10 mM Mg2+ and 100 mM NaCl (MN buffer). This increased detection sensitivity, as this combination gave a more intense band compared to MSN, at least with the selfT variant (Figure 4(c), Lane 6 vs Lane 10).
3.8. Oligomerization Study of Self Complementary Variants
Would multiple copies of the SCS allow formation of higher oligomers? Three variant tRNAsGly(GCC) were designed with multiple GGGCCC sequences. In addition, variants were designed that possessed a single SCS in either the D or anticodon loop, as distinct from the selfT variant with this sequence in the T loop. Each variant possessing a single insertion would not be expected to form higher oligomers. (The position of the SCS in these variants in relation to the cloverleaf secondary structure is shown in Figure 1(d); the variants are named to indicate the position of the SCS(s) e.g. selfD (= GGGCCC sequence in D loop). Initially a very brief interval occurred between the addition of the oligomerizing buffer and non-denaturing loading dye, and the samples were loaded immediately onto the gel. Apart from the selfT variant (+ve control), only one of the single variants, selfAc, appeared to undergo rapid oligomerization, although the selfD/Ac/T variant and (to a lesser extent) selfAc/T and selfD/Ac variants showed indications of a dimer band on an overexposed image of this gel (data not shown). To check whether these variants might undergo oligomerization over a longer time, the experiment was repeated with overnight incubation (Figure 5(a)). With this increase in incubation time, the selfD/Ac/T variant underwent dimerization along with the single selfAc variant, but apparently did not form higher oligomers above the dimer. The selfAc variant had been shown in a previous experiment to dimerize extremely quickly, and a time-course for this variant demonstrated dimerization did not depend on the components of the oligomerizing buffer and occurred even prior to renaturation; interestingly, in this experiment there was complete conversion to the dimer (Figure 5(b)).
With the exception of the selfAc variant, the RNAfold-predicted secondary structures of the self-complementary variants depart significantly from the canonical cloverleaf. This would explain why the selfD/Ac/T variant is not forming oligomers higher than dimers (Figure 5(a)). Variants with multiple self-complementary sequences have the potential to form intra-molecular interactions, as can be inferred from the structural predictions. As was the case with the selfT variant, the RNAfold secondary structure prediction for the selfAc variant suggests the SCS in the anticodon loop forms part of an alternative stem structure. In the case of the selfAc variant, the D and T loop interactions should be similar to the wild-type E. coli tRNAGly(GCC), and would presumably enforce a standard conformation on the molecule. As was the case with the selfT variant, the speed of dimerization strongly suggests that the self-complementary sequence is able to interact without structural rearrangement of the anticodon loop.
4. DISCUSSION
The aim of our study was to determine experimentally the minimum parameters for tRNA oligomerization with the cloverleaf structural template using an unmodified E. coli tRNAGly(GCC) transcript as the model tRNA. Agarose gel electrophoresis was used as a rapid screening method for the detection of dimers and higher order structures. Our initial approach was to introduce changes to the T loop in order to disrupt its interactions with the D loop and thus the central elbow region of tRNA—with the aim of freeing the two loops for interactions with other molecules. Two of the variants formed dimers, however the T- > anti-D variant was predicted to have a grossly altered conformation from the standard cloverleaf secondary structure. In contrast, the introduction of the selfcomplementary GGGCCC sequence in the selfT or selfAc variants allowed dimerization, presumably while keeping the standard tRNA conformation intact. The fast dimerization promoted by the SCS in these variants is a probable reason this sequence is found and/or causes dysfunction in known natural biological systems, such as the dimerization initiation sequence (DIS) of avian leukosis virus genomic RNA [34] and the dimer interface of the A14G mutant human mitochondrial tRNALeu(UAA) [21]. Unfortunately, additional copies of the GGGCCC sequence within the tRNA framework may not permit higher oligomers due to a preference for intramolecular interacttions between the sequences. It is likesly to be possible to construct tRNA variants that will form higher oligomers by judicious selection of complementary sequences, but whether the standard tRNA structure will be maintained with its tetragonal shape is not so certain.
4.1. Magnesium-Dependence of Oligomerization
An interesting finding of this work was the divalent magnesium and/or polyvalent spermine dependence of selfT variant oligomerization. An alternative possibility is that Mg2+/spermine stabilizes the dimers after their formation, preventing their dissociation during analysis by electrophoresis (C. Marshall, pers. commun.). The selfAc variant did not require Mg2+ for dimerization unlike the A14G mutant human mitochondrial tRNALeu(UAA), found by Kelley and colleagues to be dependent on Mg2+ (50% at 4 mM) or spermine [22]. The selfT variant by contrast in our study underwent ~70% dimerization at 1 mM Mg2+ (Figure 4(c), Lane 2). Mg2+-induced oligomerization occurred extremely quickly (Figure 4(a)), but the concentration of selfT variant transcript (5.4 µM) used in the experiment shown in Figure 7 was more than five-fold higher than in Kelley’s experiments with the A14G mutant human mitochondrial tRNALeu(UAA) (1 µM) [22]. Kelley and colleagues [22] probed the wild type and A14G mutant human mitochondrial tRNALeu(UAA) with terbium, which binds to metal-binding sites in RNA with 600 to 10,000 greater affinity than Mg2+. The bound terbium cleaves the phosphodiester backbone of tRNA, allowing for the identification of Mg2+-binding sites. They found a two-fold difference in cleavage between the wild type and A14G mutant human mitochondrial tRNALeu(UAA) at the site of the A to G mutation (the second “G” in the GGGCCC sequence); a result replicated using leadcleavage. This “suggests the formation of an additional metal-binding site at the site of the mutation [that] may stabilize the dimeric complex” [22]. The finding was
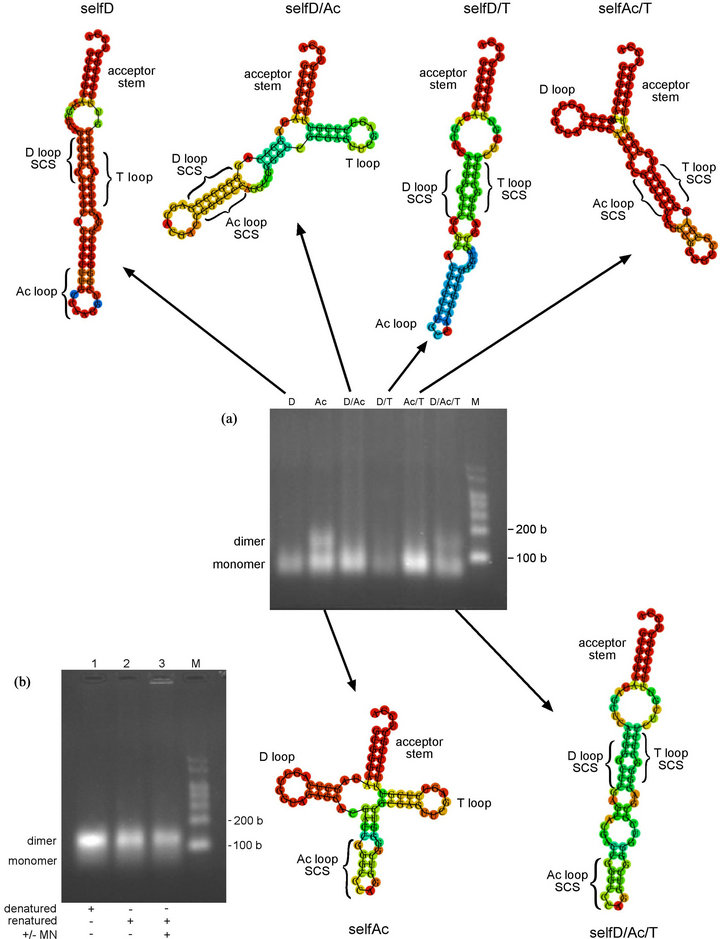
Figure 5. Oligomerization study of self-complementary variants (a) (upper) Minimum free energy (MFE) secondary structures of self-complementary variants that do not form oligomers under the conditions used. Produced using RNAfold online programme (centre) Oligomerization study of self-complementary variants in MN oligomerization buffer. Transcripts were incubated for 17 h at 4˚C. SelfD (D) (2.6 µM), selfAc (Ac) (4.9 µM), selfD/Ac (D/Ac) (4.6 µM), selfD/T (D/T) (2.8 µM), selfAc/T (Ac/T) (4.8 µM), selfD/Ac/T (D/Ac/T) (4.3 µM) (lower) Minimum free energy (MFE) secondary structures of self-complementary variants that form dimers. Produced using RNAfold online programme. SCS = self-complementary GGGCCC sequence (b) Oligomerization study of selfAc variant in absence and presence of MN oligomerization buffer using standard oligomerization protocol. SelfAc transcript at 4.1 µM. M = RNA markers. b = bases.
in agreement with the earlier work of Ennifar and colleagues [23] who solved the crystal structure of the HIV-1 subtype A dimerization initiation site (DIS) sequence, finding a Mg2+-binding site at the dimer interface between the two U275G276 dinucleotides of the paired self-complementary hexanucleotide 274-GUGCAC-279 (U275 and G276 in italics). In contrast, they found that the HIV-1 B subtype, with the self-complementary sequence 274-GCGCGC-279, did not contain a Mg2+-binding site at the corresponding position. The authors concluded that C275 did not possess the required O4 oxygen needed to coordinate the Mg2+ atom present in the A subtype [23]. This Mg2+ ion plays a crucial role, as the HIV-1 subtype A (which possesses this Mg2+-binding site) requires Mg2+ for efficient dimerization [56], in contrast to the B subtype. The Mg2+ cation may in the former case effectively minimize repulsion between phosphates on the two interacting loops (physiological Mg2+ concentration is 0.5 - 2 mM [57], so Mg2+ is not a limiting factor for dimerization). Fiore and colleagues have suggested alternatively that the Mg2+ cation may assist RNA folding by decreasing the entropic cost of counterion uptake and by reducing the disorder of the unfolded conformation [58]. In our studies, since the self-complementary sequence GGGCCC of the selfT variant is the same as that found in the A14G mutant human mitochondrial tRNALeu(UAA), it would be expected to likewise possess a Mg2+-binding site at the second G nucleotide. Unlike cytosine, guanosine has an O6 oxygen at the same relative position as U275 of the HIV-1 subtype A, which in a G-C base pair, projects into the major groove; the Mg2+ cation lies in the major groove in the A subtype HIV-1 DIS. Kelley and colleagues [22] concluded that the Mg2+-binding site of the A14G mutant human mitochondrial tRNALeu(UAA) dimer is the same as that found by Ennifar and colleagues [23] for the HIV-1 A subtype. It is likely that this is also the case here for the selfT variant dimer. Although it seems likely that the anticodon loop position of the self-complementary GGGCCC sequence in the selfAc variant of our study is optimal for forming intermolecular interactions, the lack of dependence on Mg2+- binding is difficult to understand. An online programme now exists which predicts positions of metal ions in RNA sequences [59], suggesting that the answer to some of these questions regarding the role of Mg2+ ions in RNA loop-loop interactions may become more tractable in the near future.
4.2. Possible Medical Applications of tRNA Constructs
Recent work [60] has explored the potential of using designed interacting tRNAs to rescue the disease phenotype caused by the human mitochondrial A14G tRNALeu(UAA) mutation studied by Kelley and colleagues [21,22]. Similarly, tRNAs such as the selfT variant conceivably could function as therapeutic agents to target the dimerization initiation site (DIS) of viral pathogens by an antisense mechanism. As discussed, the self-complementary GGGCCC sequence in the T loop of the selfT variant is identical to that found in the DIS of avian leukosis virus. It might be capable of blocking avian leukosis viral RNA dimerization by competitive binding, and hence virus replication in vivo (modification of other parts of the selfT variant sequence may be required to make formation of the tRNA-viral RNA heterodimer more favourable than that of the homodimers). Similarly, tRNA variants could be designed that target the DIS sequence of RNA viruses such as HIV-1. Having structural homology to endogenous tRNAs might allow such constructs to “slip beneath the radar” of the cell’s natural defenses. Proof of concept of this idea has been demonstrated by Dardel and colleagues [61,62] in work in which a tRNA “scaffold” is utilized as a means of expressing high quantities of a range of RNAs in E. coli. They found that the tRNA structure bracketing the inserted sequence is apparently sufficient to “trick” the cell into thinking that the RNA is endogenous, enabling it to escape degradation by cellular RNases.
5. ACKNOWLEDGEMENTS
Grant to WPT, and postgraduate support to HSB from Marsden Fund of NZ.
REFERENCES
- Kholod, N.S. (1999) Dimer formation by tRNAs. Biochemistry (Moscow), 64, 298-306.
- Kholod, N., Pan’kova, N., Ksenzenko, V. and Kisselev, L. (1998) Aminoacylation of tRNA transcripts is strongly affected by 3’-extended and dimeric substrate RNAs. FEBS Letters, 426, 135-139. doi:10.1016/S0014-5793(98)00320-2
- Kholod, N., Vassilenko, K., Shlyapnikov, M., Ksenzenko, V. and Kisselev, L. (1998) Preparation of active tRNA gene transcripts devoid of 3’-extended products and dimers. Nucleic Acids Research, 26, 2500-2501. doi:10.1093/nar/26.10.2500
- Moras, D., Comarmond, M.B., Fischer, J., Weiss, R. and Thierry, J.C. (1980) Crystal structure of yeast tRNAAsp. Nature, 288, 669-674. doi:10.1038/288669a0
- Westhof, E., Dumas, P. and Moras, D. (1985) Crystallographic refinement of yeast aspartic acid transfer RNA. Journal of Molecular Biology, 184, 119-145. doi:10.1016/0022-2836(85)90048-8
- Moras, D., Dock, A.-C., Dumas, P., Westhof, E., Romby, P., Ebel, J.-P. and Giegé, R. (1986) Anticodon-anticodon interaction induces conformational changes in tRNA: Yeast tRNAAsp, a model for tRNA-mRNA recognition. Proceedings of the National Academy of Sciences of the United States of America, 83, 932-936. doi:10.1073/pnas.83.4.932
- Nissen, P., Thirup, S., Kjeldgaard, M. and Nyborg, J. (1999) The crystal structure of Cys-tRNACys–EF-Tu– GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure, 7, 143-156. doi:10.1016/S0969-2126(99)80021-5
- Bénas, P., Bec, G., Keith, G., Marquet, R., Ehresmann, C., Ehresmann, B. and Dumas, P. (2000) The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA, 6, 1347-1355. doi:10.1017/S1355838200000911
- Romby, P., Giegé, R., Houssier, C. and Grosjean, H. (1985) Anticodon-anticodon interactions in solution. Studies of the self-association of yeast or Escherichia coli tRNAAsp and of their interactions with Escherichia coli tRNAVal. Journal of Molecular Biology, 184, 107-118. doi:10.1016/0022-2836(85)90047-6
- Romby, P., Westhof, E., Moras, D., Giegé, R., Houssier, C. and Grosjean, H. (1986) Studies on anticodon-anticodon interactions: Hemi-protonation of cytosines induces self-pairing through the GCC anticodon of E. coli tRNA-Gly. Journal of Biomolecular Structure and Dynamics, 4, 193-203. doi:10.1080/07391102.1986.10506339
- Gray, D.M. and Vaughan, M. (1980) Circular dichroism spectra show that repeating dinucleotide DNAs may form helices in which every other base is looped out. Nucleic Acids Research, 8, 3695-3707. doi:10.1093/nar/8.16.3695
- Mariappan, S.V., Catasti, P., Chen, X., Ratliff, R., Mazes, R.K., Bradbury, E.M. and Gupta, G. (1996) Solution structures of the individual single strands of the fragile X DNA triplets (GCC)n·(GGC)n. Nucleic Acids Research, 24, 784-792. doi:10.1093/nar/24.4.784
- Bink, H.H., Hellendoorn, K., van der Meulen, J. and Pleij, C.W. (2002) Protonation of non-Watson-Crick base pairs and encapsidation of turnip yellow mosaic virus RNA. Proceedings of the National Academy of Sciences of the United States of America, 99, 13465-13470. doi:10.1073/pnas.202287499
- Wright, H.T., Manor, P.C., Beurling, K., Karpel, R.L. and Fresco, J.R. (1979) The structure of baker’s yeast tRNAGly: A second tRNA conformation. In: Schimmel, P.R., Söll, D. and Abelson, J.N., Eds., Transfer RNA Part 1: Structure, Properties, and Recognition, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 145-160.
- Dirheimer, G., Keith, G., Dumas, P. and Westhof, E. (1995) Primary, secondary, and tertiary structures of tRNAs. In: Söll, D. and Raj Bhandary, U., Eds., tRNA: Structure, Biosynthesis, and Function, ASM Press, Washington DC, 93-126.
- Sampson, J.R. and Uhlenbeck, O.C. (1988) Biochemical and physical characterization of an unmodifed yeast phenylalanine transfer RNA transcribed in vitro. Proceedings of the National Academy of Sciences of the United States of America, 85, 1033-1037. doi:10.1073/pnas.85.4.1033
- Claesson, C., Samuelsson, T., Lustig, F. and Borén, T. (1990) Codon reading properties of an unmodified transfer RNA. FEBS Letters, 273, 173-176. doi:10.1016/0014-5793(90)81077-2
- Perret, V., Garcia, A., Puglisi, J., Grosjean, H., Ebel, J.P., Florentz, C. and Giegé, R. (1990) Conformation in solution of yeast tRNAAsp transcripts deprived of modified nucleotides. Biochimie, 72, 735-744. doi:10.1016/0300-9084(90)90158-D
- Derrick, W.B. and Horowitz, J. (1993) Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: Evidence for stable base modification-dependent conformers. Nucleic Acids Research, 21, 4948-4953. doi:10.1093/nar/21.21.4948
- Vermeulen, A., McCallum, S.A. and Pardi, A. (2005) Comparison of the global structure and dynamics of native and unmodified tRNAVal. Biochemistry, 44, 6024- 6033. doi:10.1021/bi0473399
- Wittenhagen, L.M. and Kelley, S.O. (2002) Dimerization of a pathogenic human mitochondrial tRNA. Nature Structural Biology, 9, 586-590.
- Roy, M.D., Wittenhagen, L.M. and Kelley, S.O. (2005) Structural probing of a pathogenic tRNA dimer. RNA, 11, 254-260. doi:10.1261/rna.7143305
- Ennifar, E., Walter, P., Ehresmann, B., Ehresmann, C. and Dumas, P. (2001) Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nature Structural Biology, 8, 1064- 1068. doi:10.1038/nsb727
- Yousef, M.R., Grundy, F.J. and Henkin, T.M. (2005) Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. Journal of Molecular Biology, 349, 273-287. doi:10.1016/j.jmb.2005.03.061
- Green, N.J., Grundy, F.J. and Henkin, T.M. (2010) The T box mechanism: tRNA as a regulatory molecule. FEBS Letters, 584, 318-324. doi:10.1016/j.febslet.2009.11.056
- Feinberg, J.S. and Joseph, S. (2006) A conserved basepair between tRNA and 23S rRNA in the peptidyl transferase center is important for peptide release. Journal of Molecular Biology, 364, 1010-1020. doi:10.1016/j.jmb.2006.09.040
- Kim, D.F. and Green, R. (1999) Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Molecular Cell, 4, 859-864. doi:10.1016/S1097-2765(00)80395-0
- Busch, S., Kirsebom, L.A., Notbohm, H. and Hartmann, R.K. (2000) Differential role of the intermolecular base-pairs G292-C75 and G293-C74 in the reaction catalyzed by Escherichia coli RNase P RNA. Journal of Molecular Biology, 299, 941-951. doi:10.1006/jmbi.2000.3789
- Fauzi, H., Jack, K.D. and Hines, J.V. (2005) In vitro selection to identitfy determinants in tRNA for Bacillus subtilis tyrS T box antiterminator mRNA binding. Nucleic Acids Research, 33, 2595-2602. doi:10.1093/nar/gki546
- Gutiérrez-Preciado, A., Yanofsky, C. and Merino, E. (2007) Comparison of tryptophan biosynthetic operon regulation in different Gram-positive bacterial species. Trends in Genetics, 23, 422-426. doi:10.1016/j.tig.2007.05.005
- Marck, C. and Grosjean, H. (2002) tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA, 8, 1189-1232. doi:10.1017/S1355838202022021
- Scarabino, D., Crisari, A., Lorenzini, S., Williams, K. and Tocchini-Valentini, G.P. (1999) tRNA prefers to kiss. EMBO Journal, 18, 4571-4578. doi:10.1093/emboj/18.16.4571
- Song, R., Kafaie, J., Yang, L. and Laughrea, M. (2007) HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. Journal of Molecular Biology, 371, 1084-1098. doi:10.1016/j.jmb.2007.06.010
- Polge, E., Darlix, J.-L., Paoletti, J. and Fossé, P. (2000) Characterization of loose and tight dimer forms of avian leukosis virus RNA. Journal of Molecular Biology, 300, 41-56. doi:10.1006/jmbi.2000.3832
- Horiya, S., Li, X., Kawai, G., Saito, R., Katoh, A., Kobayashi, K. and Harada, K. (2002) RNA LEGO: Magnesium-dependent assembly of RNA building blocks through loop-loop interactions. Nucleic Acids Research Supplement, 2, 41-42. doi:10.1093/nass/2.1.41
- Horiya, S., Li, X., Kawai, G., Saito, R., Katoh, A., Kobayashi, K. and Harada, K. (2003) RNA LEGO: Magnesium-dependent formation of specific RNA assemblies through kissing interactions. Chemistry & Biology, 10, 645-654. doi:10.1016/S1074-5521(03)00146-7
- Fujiya, I. and Harada, K. (2004) Formation of dendrimeric RNA assemblies through RNA loop-loop interacttions. Nucleic Acids Symposium Series, 48, 97-98. doi:10.1093/nass/48.1.97
- Severcan, I., Geary, C., Verzemnieks, E., Chworos, A. and Jaeger, L. (2009) Square-shaped RNA particles from different RNA folds. Nano Letters, 9, 1270-1277. doi:10.1021/nl900261h
- Severcan, I., Geary, C., Chworos, A., Voss, N., Javocetty, E. and Jaeger, L. (2010) A polyhedron made of tRNA. Nature Chemistry, 2, 772-779. doi:10.1038/nchem.733
- Hofacker, I.L., Fontana, W., Stadler, P.F., Bonhoeffer, S., Tacker, M. and Schuster, P. (1994) Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie, 125, 167-188. doi:10.1007/BF00818163
- Zuker, M. and Stiegler, P. (1981) Optimal computer folding of large RNA sequences using thermodynamic and auxiliary information. Nucleic Acids Research, 9, 133-148. doi:10.1093/nar/9.1.133
- McCaskill, J.S. (1990) The equilibrium partition function and base pair binding probabilities for RNA secondary structures. Biopolymers, 29, 1105-1119. doi:10.1002/bip.360290621
- Bernhardt, H.S. and Tate, W.P. (2006) Transfer RNA as a potential building block for nanotechnology. Chemistry in New Zealand Journal, 70, 7-11.
- Laughrea, M. and Jetté, L. (1996) Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry, 35, 1589-1598. doi:10.1021/bi951838f
- Bernhardt, H.S. and Tate, W.P. (2008) Self-assembling transfer RNA has potential for nanoparticle arrays. Current Applied Physics, 8, 380-382. doi:10.1016/j.cap.2007.10.035
- Durant, P.C. and Davis, D.R. (1999) Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. Journal of Molecular Biology, 285, 115-131. doi:10.1006/jmbi.1998.2297
- Bénas, P., Bec, G., Keith, G., Marquet, R., Ehresmann, C., Ehresmann, B. and Dumas, P. (2000) The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA, 6, 1347-1355. doi:10.1017/S1355838200000911
- Cabello-Villegas, J., Winkler, M.E. and Nikonowicz, E.P. (2002) Solution conformations of unmodified and A37N6- dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. Journal of Molecular Biology, 319, 1015-1034. doi:10.1016/S0022-2836(02)00382-0
- Kim, C.-H. and Tinoco Jr., I. (2000) A retroviral RNA kissing complex containing only two G·C base pairs. Proceedings of the National Academy of Sciences of the United States of America, 97, 9396-9401. doi:10.1073/pnas.170283697
- Lagerkvist, U. (1978) “Two out of three”: An alternative method for codon reading. Proceedings of the National Academy of Sciences of the United States of America, 75, 1759-1762. doi:10.1073/pnas.75.4.1759
- Lagerkvist, U. (1981) Unorthodox codon reading and the evolution of the genetic code. Cell, 23, 305-306. doi:10.1016/0092-8674(81)90124-0
- Samuelsson, T., Axberg, T., Borén, T. and Lagerkvist, U. (1983) Unconventional reading of the glycine codons. Journal of Biological Chemistry, 258, 13178-13184.
- Claesson, C., Samuelsson, T., Lustig, F. and Borén, T. (1990) Codon reading properties of an unmodified transfer RNA. FEBS Letters, 273, 173-176. doi:10.1016/0014-5793(90)81077-2
- Chu, W., Liu, J.C. and Horowitz, J. (1997) Localization of the major ethidium bromide binding site on tRNA. Nucleic Acids Research, 25, 3944-3949. doi:10.1093/nar/25.19.3944
- Quigley, G.J., Teeter, M.M. and Rich, A. (1978) Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proceedings of the National Academy of Sciences of the United States of America, 75, 64-68. doi:10.1073/pnas.75.1.64
- Jossinet, F., Paillart, J.C., Westhof, E., Hermann, T., Skripkin, E., Lodmell, J.S., Ehresmann, C., Ehresmann, B. and Marquet, R. (1999) Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA, 5, 1222-1234. doi:10.1017/S1355838299990982
- Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K. and Watson, J.D. (1994) Molecular biology of the cell. 3rd Edition, Garland Publishing, New York.
- Fiore, J.L., Holmstrom, E.D. and Nesbitt, D.J. (2012) Entropic origin of Mg2+-facilitated RNA folding. Proceedings of the National Academy of Sciences of the United States of America, 109, 2902-2907. doi:10.1073/pnas.1114859109
- Philips, A., Milanowska, K., Lach, G., Boniecki, M., Rother, K. and Bujnicki, J.M. (2012) MetalionRNA: Computational predictor of metal-binding sites in RNA structures. Bioinformatics, 28, 198-205. doi:10.1093/bioinformatics/btr636
- [61] Karicheva, O.Z., Kolesnikova, O.A., Schirtz, T., Vysokikh, M.Y., Mager-Heckel, A.M., Lombès, A., Boucheham, A., Krasheninnikov, I.A., Martin, R.P., Entelis, N. and Tarassov, I. (2011) Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic Acids Research, 39, 8173-8186. doi:10.1093/nar/gkr546
- [62] Ponchon, L. and Dardel, F. (2007) Recombinant RNA technology: The tRNA scaffold. Nature Methods, 4, 571- 576. doi:10.1038/nmeth1058
- [63] Ponchon, L., Beauvaism, G., Nonin-Lecomtem, S. and Dardel, F. (2009) A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nature Protocols, 4, 947-959. doi:10.1038/nprot.2009.67
NOTES
*Corresponding author.

