Advances in Bioscience and Biotechnology
Vol.3 No.2(2012), Article ID:18449,8 pages DOI:10.4236/abb.2012.32016
Polymorphism identification of carotenoid binding protein gene transcription in the silkworm, Bombyx mori
![]()
National Engineering Laboratory for Modern Silk, Department of Applied Biology, Medical College, Soochow University, Suzhou, China
Email: *szsqxu@suda.edu.cn, myaynys@163.com, longjing_520@yahoo.cn, simyh@suda.edu.cn
Received 1 January 2012; revised 16 February 2012; accepted 10 March 2012
Keywords: Carotenoid-Binding Protein Gene; Alternative Splicing Transcripts; Single Nucleotide Polymorphism; Bombyx mori
ABSTRACT
Carotenoids play important and diverse roles in insects and their uptake and transport rely on carotenoid binding protein (CBP). The study excavated a cluster of CBP-like transcripts, including full CBP from all of the six yellow cocoon Bombyx strains investigated. Sequencing of 54 cDNA clones revealed 17 different types of transcripts which derived from alternative splicing of CBP gene locus. Five of the novel transcripts were similar with spatial and temporal distribution patterns to CBP, but their expression levels were relatively lower. The author disclosed two more novel alternative spliced transcripts with different transcription start sites from CBP in the 5’ UTRs as well as 11 SNP sites neighboring intron 1 after amplification and sequencing. qRT-PCR analysis gave evidence that relatively more mRNA was transcribed from A-type CBP gene than that from B-type in tissues like silk gland and midgut. Sequences of Aand B-type CBP genes were different in length of domains neighboring the 5’ UTR, thus their mRNA varied both in quantity and transcript types. The SNPs surrounding intron 1 can serve as stable markers to distinguish transcripts from the two isoforms, and they can be used for molecular marker assisted selection.
1. INTRODUCTION
Carotenoids play important and diverse roles in insects, and they are essential nutrients for most of insect species, even for human beings. The possible role for carotenoids in disease prevention of common chronic human diseases such as cancer, age-related macular degeneration and cardiovascular diseases has stimulated wide interest in carotenoid absorption and metabolism [1].
However, mechanisms for the uptake and transport of carotenoids are not well understood in any animal system. The domestic silkworm (Bombyx mori L.), an important economic species, is proposed as an animal model to elucidate this mechanism benefited from the research progress of carotenoid binding protein (CBP) [2,3] .
Carotenoids are supposed to be the major pigment contributing to yellow cocoon coloration, and the uptake of carotenoids into the intestinal mucosa and the silk gland is controled by the Y (Yellow blood) gene in Bombyx mori [4]. A carotenoid binding protein (CBP) was isolated from the silk gland of the fifth instar Bombyx larvae [2]. CBP can be only found in Y gene dominant strains (Y/Y) with yellow cocoons rather than recessive strains (+Y/+Y) with white cocoons [3-6]. RFLP analysis of the genes showed that CBP and the Y-gene were at the same chromosome locus and they may correspond to each other [7]. Sakudoh and colleagues provided evidence that the Y gene corresponds to the intracellular CBP gene, and demonstrated the generation of a silkworm strain with colored silk by germ-line transformation of the CBP gene into a white cocoon strain [3].
Based on the CBP gene sequences and their expression patterns in white cocoon strains and yellow cocoon strains, it was speculated that targeted insertion of a transposable element (CBP associated TRAS-like sequence, CATS) to intron 2 of CBP gene caused genome reshuffle in the neighboring area [3,5]. As a result, part of the coding sequence in CBP was deleted, making carotenoid absorption seriously disrupted and white cocoon thus generated. According to these studies, CBP gene had been classified into three types, corresponding to Y-a (A), Y-b (B) and Y-c (C) respectively. A-type only lacked the insertion of full CATS in intron 2 compared with B-structure. C-type had a deletion of about 4.5 kb between exon 2 and exon 3 and retained partial CATS [3]. Sequence composition beyond these reshuffled domains was postulated to be the same among different gene structures. Afterwards, a unique CBP gene structure (designated as D-type) was identified from the well-accepted genome sequencing strain Dazao (p50T) [8,9]. Like white cocoon strains, Dazao strain could not absorb carotenoids possibly due to the lack of CBP production [9,10], although the cocoon color was green.
In this article, we present evidence to support the existence of both sequence and functional differences between Aand B-type CBP genes, and look for reliable sequences to distinguish the relative transcription level of CBP from different gene structures in the same tissue.
2. MATERIALS AND METHODS
2.1. Experimental Animals
Multiple strains of Bombyx mori with various cocoon colors were used. In these strains, N4, MG, Y12m, Y23, Y12b and Ysf generate yellow cocoons, strain C108 generate white cocoons, while strains of p50, G1 and Gjv produce green cocoons. All strains are normally cultured in Soochow University and reared on fresh mulberry leaves under a 16/8 (light/dark) period at 25˚C.
2.2. RNA isolation and RT-PCR Analysis
Total RNA was extracted from larvae of Bombyx mori (n = 3 - 7) using TRizol Reagent (TAKARA, Japan). 2 μg RNA was mixed with 500 ng Oligo (dT)18 and heated at 70˚C for 10 min. After chilling on ice for 30 s, RNA was transcribed at 42˚C for 1 h using 500 units RevertAidTM M-MuLV Reverse Transcriptase (Fermentas, Canada).
1 μL of the cDNA product and 0.2 unit of Taq polymerase (Takara, Japan) were used in RT-PCR analysis. The conditions were as follows: 2 min at 94˚C; 30 s at 94˚C, 40 s at 54˚C and 2 min at 72˚C for 28 cycles; and 8 min at 72˚C. For CBP amplification primer C-exR3 (5’-CGGATGAGGCGGTTGTGTGCGCGTT-3’) and CexR4 (5’-AACCTGGCGGGACAGTTGACGACGC-3’) were used along with another pair C-exR1 (5’-GCGATGTGTAGCTCCGTG-3’) and C-exR2 (5’-GCCGGATGTCCTAGTGGT-3’).
Expression analysis of novel CBP transcripts were carried out using primers CR31-S (5’- CGCGACTCGAACTTCGGA-3’) and CR42-S (5’-CGCAGAAAACACTTAGAGATAGTCA-3’) with C-exR2. Actin transcripts were amplified as an external control with primer Actin-1 (5’-CCCCATCGAACACGGAATCG-3’) and Actin-2 (5’-CGCTCGGCAGTGGTAGTGAA- 3’).
2.3. RT-qPCR Analysis
The same cDNA samples were used as a template after diluting 100 times. Two pairs of primers were used in quantitative RT-PCR analysis containing SNP sites underlined, CAS (5’-GATGTGTAGCTCCGTGTGTCGTAA-3’) and CAT (5’-CTCGATTGCGGGTACGTAACTAT-3’) for A-type CBP gene amplification, CBS (5’- GATGTGTAGCTCCGTGCGTTGTAA-3’) and CBT (5’- GCTTGATTGCGGGTACGTAACTA-3’) for B-type CBP gene amplification. Fluorescent-based Realtime PCR was carried out with the ABI 7300 Real Time PCR system. SYBR Green Kit was used according to the manufacturer’s instruction (Takara, Japan). The conditions for PCR reactions were as followed: initial denaturizing at 95˚C for 1 min followed by 45 cycles of denaturizing at 95˚C for 15 s, annealing at 60˚C for 30 s. CBP transcripts’ level was normalized with Actin transcript level in the same sample.
2.4. Rapid Amplification of 5’ cDNA End
2 μg total RNA from the fifth instar larval silk glands of N4 strain was used as an initial template. Subsequent procedures were carried out using a 5’ Full RACE kit (TAKARA, Japan) to amplify 5’ cDNA end of CBP mRNA. Outer primer (5’-CATGGCTACATGCTGACAGCCTA-3’) and inner primer (5’-CGCGGATCCACAGCCTACTGATGATCAGTCGATG-3’) were used with primer C-ex3 (5’-CCGTAAATCTATAAACCTTGCCGAA-3’) in PCR analysis.
2.5. Cloning and Sequencing
PCR products were separated by running on a 1.2% agarose gel under 120 V and visualized using EtBr staining method. Bands were separately cut out or purified in mixture. The purified fragments were ligated with pMD19-T vector (TAKARA, Japan) and further transformed into competent Escherichia coli cells. Cells were cultured on LB agar containing ampicillin (100 μg/mL). Positive clones were screened by restrictive reactions with HindШ and XbaI. Sequencing was done by BGI in Shanghai, China.
2.6. Sequence Analysis
CBP gene sequences were extracted from GenBank by NCBI Blast tool under accession numbers: AB211536, AB211535, AB263197-AB263200, AB062740, BABH- 01017793. Sequence alignment was done by Clustal W [11].
3. RESULTS
3.1. Transcription Polymorphism of CBP in Different Strains
Initially, CBP expression was investigated in Bombyx strains with varied cocoon colors. In white cocoon strain like C108 and green cocoon strain like p50, a single band (406 bp) was amplified and further confirmed to be lack of exon 2. And a single band of 715 bp was detected in yellow cocoon strains under the same experimental conditions. However, multiple bands were observed and the truncated CBP form lacking exon 2 was also amplified (see Figure 1(a)) in accordance with our previous report [8]. Similar results were obtained using another primer pair covering the whole ORF region of CBP (Figure 1(b)), suggesting that these bands didn’t derive from amplification by mutations in the primer annealing sites. A preliminary sequencing and sequence alignment were carried out. The result showed that these bands were highly similar to CBP and they differed mainly in the 5’ part (Figure 1(b)). Thus, they were named CBP-like transcripts featuring their length polymorphism. Compared with CBP gene sequence [3,5], the exon-intron boundaries of these transcripts all conform to the “GT/AG” rule
 (a)
(a)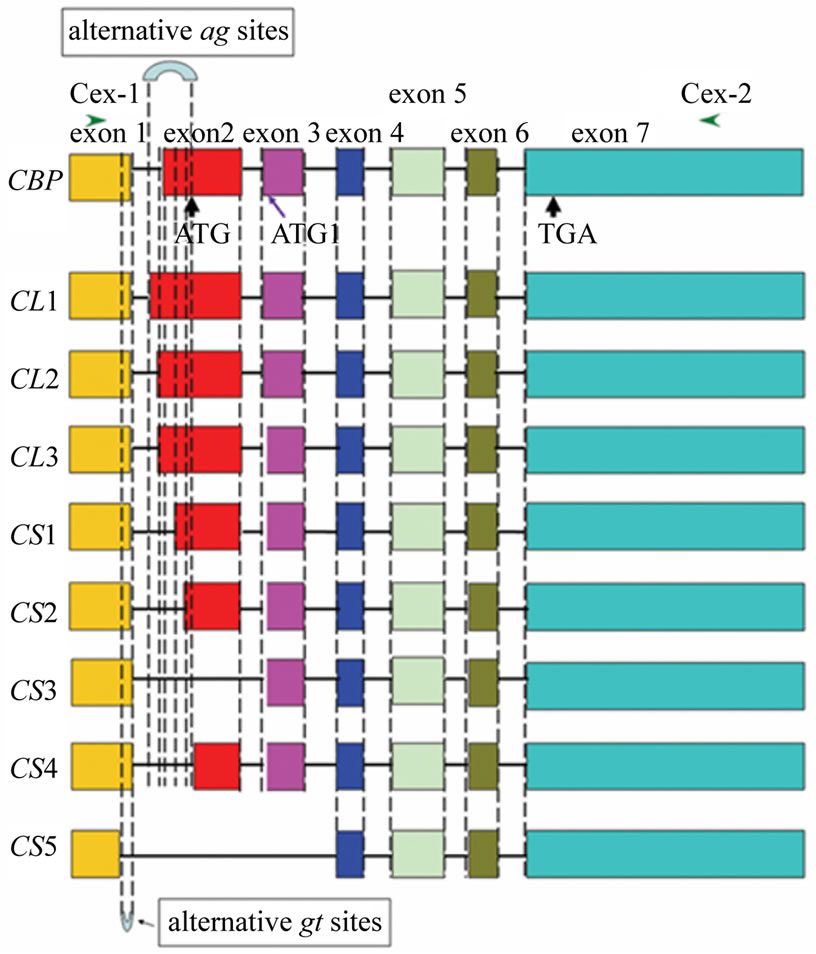 (b)
(b)
Figure 1. Investigation of CBP expression in Bombyx strains with varied cocoon colors (a) and alignment of alternative spliced transcripts with full CBP sequence (b). (a) Primers C-exR3 and C-exR4 were used in RT-PCR analysis for CBP. The expected band size was 715 bp in Bombyx strains with yellow cocoons like N4 and 406 bp for the other colored-cocoon strains; (b) Primer pairs C-ex1 and C-ex2 were used to obtain fragments covering the whole ORF. Box with varied grayscales indicated each exon and lines for introns. The intron length was not drawn to scale. Start (or alternative start) codon and stop codon were arrow-headed. All exon and intron boundaries conformed to “GT/AT” which was represented by dashed vertical lines as indicated in the figure.
[12], suggesting they originate from alternative splicing events.
3.2. Identification of CBP-Like Transcripts in N4
In this study, strain N4 was chosen to investigate all possible forms of CBP-like transcripts. In the result, a total number of 17 CBP-like transcripts were identified after sequencing 54 cDNA clones. More than one forth of these cDNA clones corresponded to full CBP (Figure 2). Five cDNA clones (9%) contained enlarged sequence of CBP-like transcripts. Truncated forms of CBP were found in the remaining cDNA clones (65%). Almost 80% of these transcripts varied in sequence between exon 1 and exon 2 (Figure 2). These transcripts all conform to “GT/ AG” rule in regard to the truncated or integrated sequence boundaries. A BmStart1 gene was previously identified as a transcription variant of CBP [5], suggesting alternative transcripts may not be rare in this gene locus.
3.3. Novel CBP Transcripts Identified by 5’ RACE Analysis
To verify the existence of other forms of alternative transcripts, rapid amplification of cDNA end (RACE) was employed to obtain 5’ UTR of CBP. As a result, another thirteen distinctive transcripts were identified besides the complete CBP form. These distinctive products thus were called novel CBP transcripts to differentiate

Figure 2. Classification of CBP-like transcripts. Primers CexR1 and C-exR2 were used to amplify area covering the whole CDS region of CBP and a total number of 54 cDNA clones were sequenced and aligned. (a) Transcripts were mainly classified into three groups according to their length related to the complete CBP form; (b) Transcripts showing variation in each exon.
themselves from CBP-like transcripts. They were also alternatively spliced and could be classified into two groups according to transcription start site (TSS), i.e. CBP variant 31 (CR31) and CBP variant 42 (CR42). CR31 series transcripts were found to start from the upstream of CBP gene and CR42 initiated in intron 2. After exploring the NCBI EST database of Bombyx mori by “Blast” method, two EST tags were found highly similar with CR31 transcripts by accession numbers BB985640 and BB986255, but CR42 transcripts corresponded to unknown EST tags.
Sequence alignment showed that some CR31 transcripts even contained fragment of intron 1 sequence. Thus, special primers located at 5’ end of novel transcripts and 3’ end of CBP respectively, which would amplify most of the conserved area spanning from exon 3 to exon 7 of CBP, were designed to test whether any form of CR31 or CR42 was truly expressed. RT-PCR and subsequent sequencing analysis identified three forms of CR31 mRNA and two of CR42. Tissue distribution pattern of CR31 mRNA products were similar to that of CBP, while CR42 mRNA was expressed mainly in the gonad (see Figure 3). Similar to CBP expression patterns [10,13], expression of these transcripts was repressed in silk gland during the fourth instar (data not shown), suggesting a common mechanism of expression regulation on these genes and thus giving this special spatial-temporal expression pattern.
3.4. Discrimination of Afrom B-Type CBP Transcripts via SNPs in the Regions Neighboring Intron 1
Previously we reported an intron 1 variant (intron 1v) of CBP gene [8]. It was distinctive both in sequence and length (1225 bp) compared with its counterpart (1541 bp)
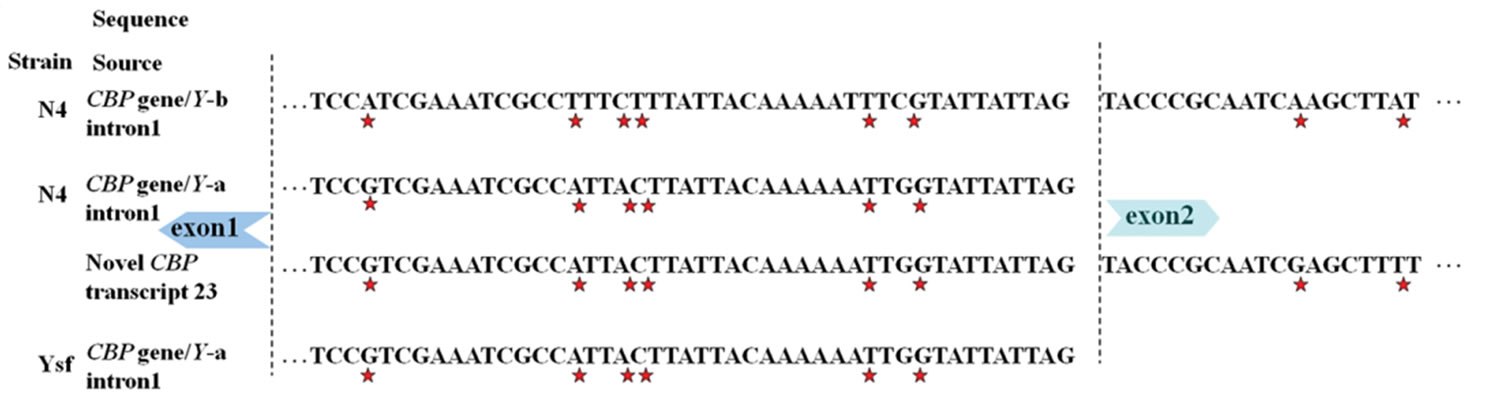
in B/C-type CBP gene [3]. For this result, intron 1v was found to exist only in yellow cocoon strains of Bombyx mori under investigation, and we speculate it as a unique sequence composition of A-type CBP gene. Alignment was carried out for the two distinctive intron sequences and at least 7 SNP sites were identified in the proximate 3’ end. Furthermore, 3’ end region of intron 1v, rather than intron 1 was detected in a CBP-like transcript (see Figure 4). Sequence inclusion of 5’ end of intron 1v was also observed in some CBP-like transcripts.
We also discovered 8 SNPs in the partial sequence of exon 1 and exon 2 from PCR experiment above for intron 1 amplification and sequence analysis. This led us to further align all 14 full CBP sequences from cDNA cloning experiment above. The 8 same SNPs did appear and another 3 SNPs were identified in the 5’ UTR (see Table 1). Therefore these SNPs were considered to be the additional proof of sequence difference between Aand B-type CBP gene. A length polymorphism of consecutive deoxyadenosine nucleotides was also found which lied at –23 from the start codon. There were 10 and 7 consecutive adenine nucleotides in Aand B-type CBP sequence respectively.
3.5. CBP Gene Expression Analysis
Gene structure difference may result in expression variation. We strived to examine whether Aand B-type CBP gene differed in expression level. Quantitative RT-PCR analysis was carried out according to the expression of sorbitol dehydrogenase-2a and 2b genes, which shared most of their sequence [14]. Two primer pairs with SNPs in the 5’ UTR were designed according to Aand B-type CBP gene sequence identified in this study. Consequently, mRNA expression level of A-type CBP was found al-
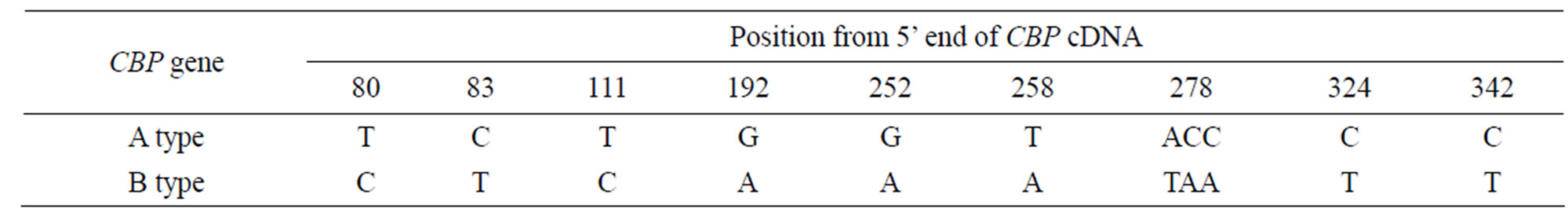
Table 1. SNPs identified in 5’ UTR of CBP gene.
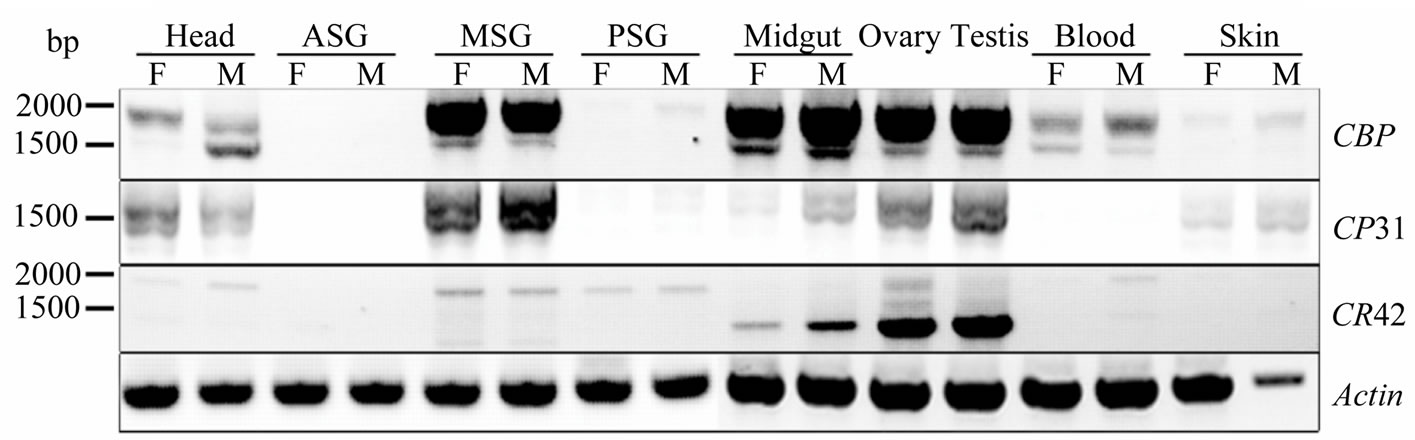
Figure 3. Tissue distribution pattern of CBP and novel transcripts. Individuals (5 - 7) of day 3 of the fifth instar larvae were used to extract total RNA. CBP was amplified with primers CexR1 and C-exR2.
 (a)
(a) (b)
(b)
Figure 4. Distinction of A type from B type CBP gene via SNPs. Arrowhead indicated the start codon of carotenoid binding protein (CBP) gene. (a) SNP sites identified in part of intron 1 and neighboring exon 2; (b) SNPs near the start codon and a consecutive adenine nucleotide length polymorphism. Other sequences with accession numbers were extracted from published data [2,5,6]. Stars under nucleotides or plus sign up nucleotides marked SNPs.
most the same as B-type CBP in the midgut from female individuals. In other tissues like ovary, mRNA level of A-type CBP was 4.9% - 26.2% higher than B-type mRNA (Figure 5). Also, CBP expression level in male tissues was often higher than that in female counterparts. Since Y-a type CBP gene was disclosed to exist as a single copy, and Y-b had three copies in strain N4 [9], it is worth investigating whether transcription activity of these two genes are actually different or not.
4. DISCUSSION
4.1. Alternative Splicing May Be Responsible for CBP Gene Transcription Polymorphism
Alternative splicing (AS) in Bombyx mori is not rare, and an alternative spliced CBP isoform called BmStart1 had been reported [5]. It is common in yeast [15], human beings [16,17] and other organisms. The nicotinic acetylcholine receptor (nAChRs) gene family in insects [18] and Bombyx mori [19] can bear alternative splicing activities, and the GC content is very important in this AS process. Schwartz and colleagues found nucleosome occupancy within exons was closely correlated with GC content, with the highest occupancy level in intermediate GC content of 41% - 57%, thus favoring exon inclusion in mRNA [20]. Human exons tend to be GC-rich when compared with the intronic regions adjacent [21].
In this study, CBP gene transcribed a cluster of related transcripts with length polymorphism in yellow cocoon strains of Bombyx mori. Other novel CBP transcripts were also identified with no strain limit according to cocoon colors. They paralleled in conservation with CBP, including the same or most part of the coding sequence. Since AS recognition sites existed in CBP gene, the GC content of exon and alternatively spliced intron may fluctuate. GC contents were investigated in domains surrounding intron 1 where most sequence variants appeared in CBP gene. As a result, the GC contents of exon 1 (36.7% - 47.8%) and exon 2 (41.4% - 47.0%) in all transcripts were actually higher than those of intron 1 (35.2% - 36.3%) (data not shown).
4.2. CBP Gene Transcription Polymorphism Varied by Cocoon Colors and Existed in Each Larval Stage
Wakamatsu et al. pointed that one gene produces suitable protein-coding transcripts responding to the situation and environment [22]. In our data, the cDNA clones for
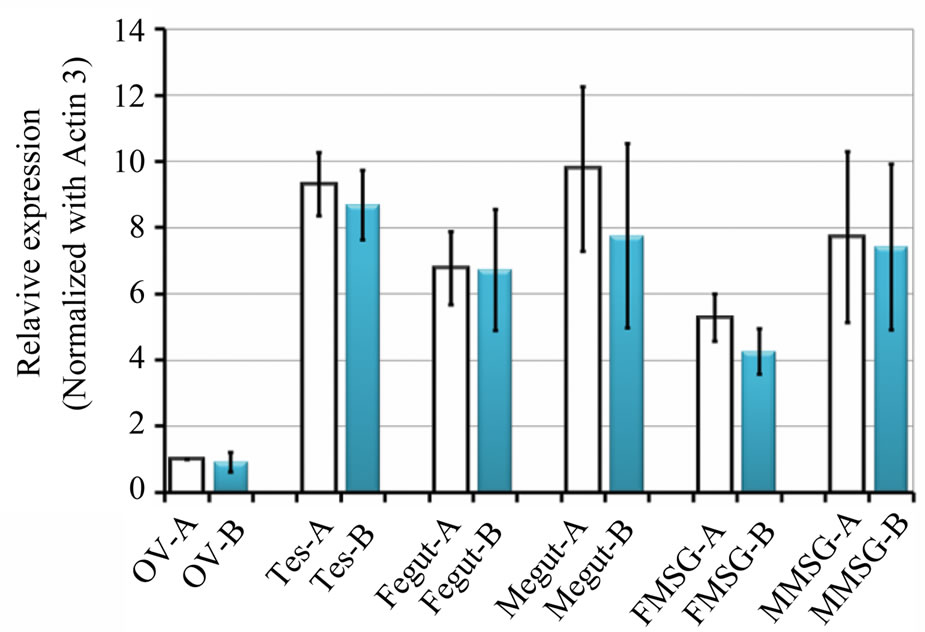
Figure 5. Quantitative expression analyses of Aand B-type CBP gene. The same samples were used with two different primer pairs containing SNPs (see Materials and Methods). Vertical axis indicated the fold increase of CBP mRNA compared to A-type mRNA in ovary. Each value represented an average fold increase with standard errors (n = 3). Ov, ovary; Tes, testis; Fegut and Megut, midgut from female and male individuals; FMSG and MMSG, middle silk gland from female and male individuals. Bars stand for standard derivatives.
transcript investigation were obtained from multiple tissues with high CBP expression including silk glandmidgut, testis and ovary. Therefore tissue-specific transcription may produce different kinds of CBP-like transcripts. Among these transcripts, some of them have full coding sequence for CBP. Thus it is necessary to investigate whether these transcripts are tissue-specific. The transcription polymorphism happened mainly in yellow cocoon strains of Bombyx mori containing multiple copies of CBP gene, which varied especially by cocoon colors. We also found that CBP gene transcription polymorphism existed in each larval stage.
4.3. A-Type Differed from B-Type CBP Gene Not Only in Absence of a Transposable Element Called CATS
Besides the well-characterized A/B/C-type CBP gene sequence difference [3], a unique D-type CBP gene in Dazao was identified [8,9], which was found substantially different from A/B/C-type. There was no exon 2 or intron 2, but two new intron sequences lie downstream of exon 1 and exon 3 accordingly. To objectify the structural differences among these four CBP types, we redrew them for comparison using published sequence data (Figure 6(b)) [2,3,5,8,23].
 (a)
(a)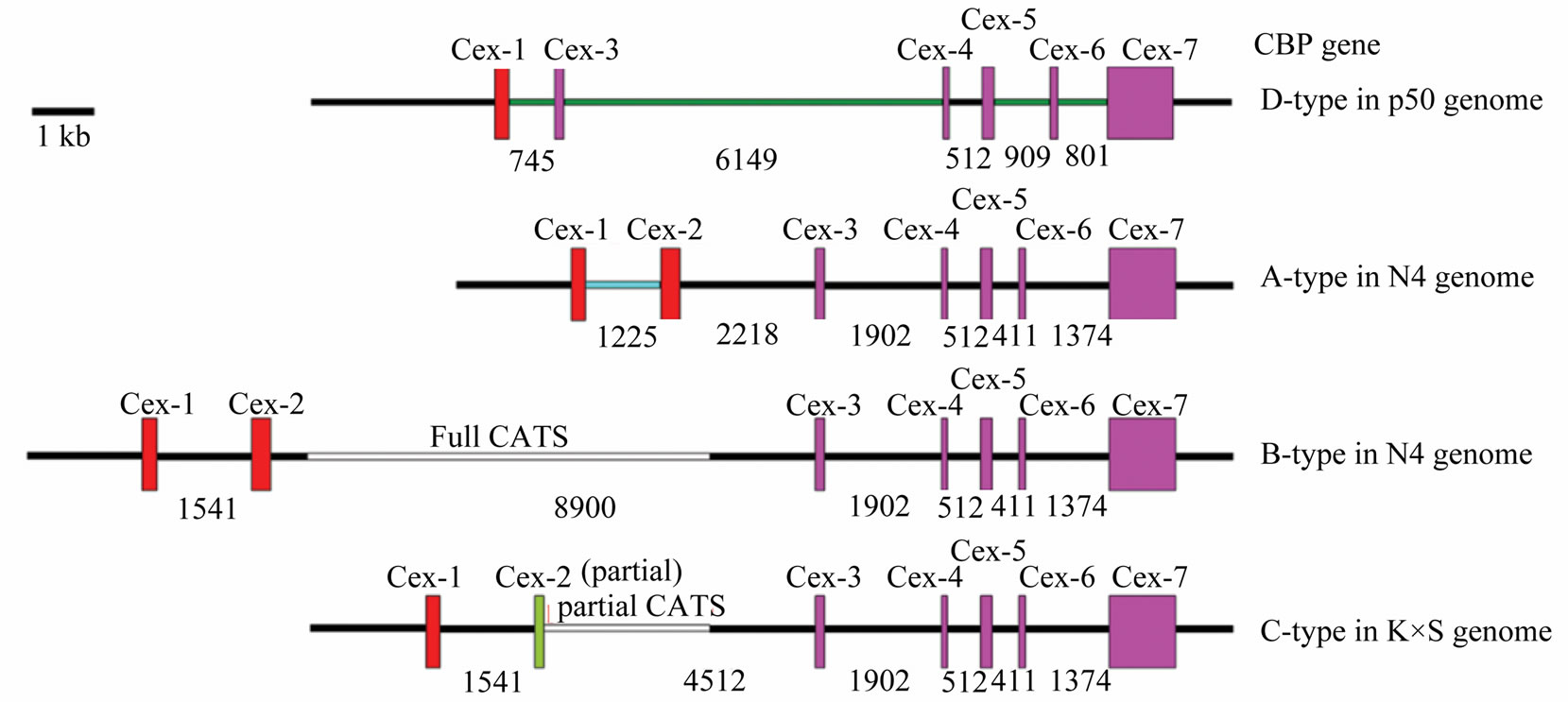 (b)
(b)
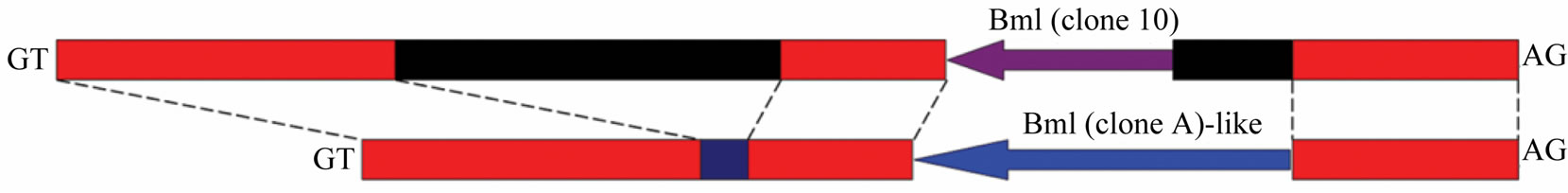
Figure 6. Detailed intron 1 comparison and different structures of CBP gene according to strains of Bombyx mori. (a) Bm1 repetitive DNA element was identified in intron 1 and intron 1v. The element in intron 1v was 50 bp shorter in 3’ end than Bm1 (clone-A) sequence. Red boxes indicated highly similar sequence, black for intron 1 specific sequence, and blue for intron 1v unique sequence; (b) Detailed structures of CBP gene according to Bombyx strains. Exons were shown as box and inton drawn in line; numbers indicated intron length. Note: each CBP gene structure has a different intron 1 sequence drawn with colored lines, except that Band C-type share the same intron 1. Exon 2 and intron 2 were also different among structures. Gene structures were drawn according to genomic information of Bombyx mori in GenBank by accession no. AB211535, AB263200, BABH01017793 and GQ847750.
It has been hypothesized that Adiffered from B-type CBP gene in absence of a transposable element called CATS [5]. Our experimental results showed that there should be other sequence differences.
First of all, A-type CBP gene contains a distinctive intron 1v, rather than the published intron 1 shared by both Band C-type CBP gene. We had previously reported preliminary sequence alignment of intron 1 and intron 1v [8], and the two intron sequences contained fragment highly similar with some expressed tags (ESTs). A further nt/nr database search identified three different Bm1 (sequence involved in CATS insertion to intron 2) repetitive DNA elements [5,24], namely clone-10 in intron 1 and clone-A in intron 1v and clone-10 in the exon 7 of CBP gene. The clone-A element was longer in the 5’ end than that of clone-10.
Second, SNPs were identified. Previously we had compared CBP gene sequences extracted from published data where 17 SNPs were identified in the putative ORF region. In this paper, the existence of these SNPs was proved again via comparing sequencing data of the truncated CBP from Dazao strain with full CBP isolated from strain N4. The truncated CBP from white and green cocoon strains of Bombyx mori were also separately isolated and sequenced, but no SNP was found in the 5’ UTR except in Dazao strain.
4.4. A-Type CBP Gene Produce More mRNA than That of B-Type Gene
The rearing of natural colored cocoon silkworm is being more and more applied in main sericulture countries such as China, Thailand and India, benefitting from that the cocoon can been used to produce chemical-stain-free textile. Notwithstanding the great advantages, we are faced with issues in color controlment and shorter breeding cycle. Generally, it is urgent to come up with effective molecular marker assisted selection.
Via SNPs, 10 in 14 cDNA clones containing full length CBP were classified as A-type CBP gene products and the others as B-type products. The majority (90%) of those CBP-like transcripts were also from A-type CBP gene transcription. For novel CBP transcripts, over 90% were identified as A-type CBP gene transcripts (Table 2). These results showed transcription polymorphism of CBP gene locus. qRT-PCR analysis showed that A-type CBP gene could actually produce more mRNA than that of B-type gene. Male and female expression disparity of these two genes was also detected in the examined tissues.
In the future research, the relative individual expression level of these two CBP gene types in Bombyx mori will be assessed, hoping to give a more detailed expression pattern influenced by gene structure disparity.
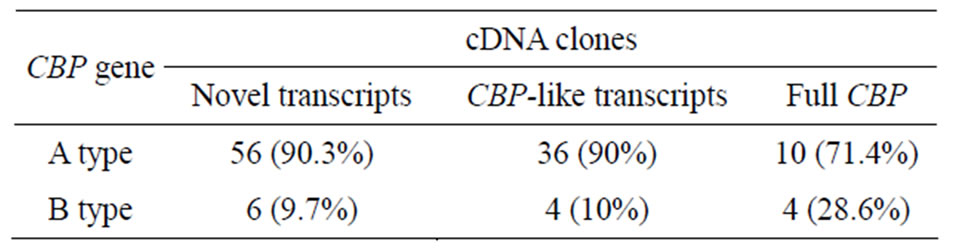
Table 2. Transcripts identified as of Aor B-type CBP gene products.
4.5. Nucleotide Sequence
Nucleotide sequences identified in this paper have been deposited in GenBank under accession numbers GU32- 4982, HM165191.
5. ACKNOWLEDGEMENTS
We thank Ms Xuemei Duan for her technical support. This work was supported by the National High-tech R&D Program of China (863 Program) (Grant No. 2011AA100306), the Priority Academic Program Development of Jiangsu Higher Education Institutions, National Natural Science Foundation of China (Grant No. 31172264), Major Applied Research Program of Soochow University (Project No. Q3034850).
REFERENCES
- Mayne, S.T. (1996) Beta-carotene, carotenoids, and disease prevention in humans. Official Publication of the Federation of American Societies for Experimental Biology, 10, 690-701. doi:10.1096/fj.08-125435
- Tabunoki, H., Sugiyama, H., Tanaka, Y., Fujii, H., Banno, Y., Jouni, Z.E., Kobayashi, M., Maekawa, H. and Tsuchida, K. (2002) Characterization, and cDNA sequence of a carotenoid binding protein from the silk gland of Bombyx mori larvae. Journal of Biological Chemistry, 277, 32133-32140. doi:10.1074/jbc.M204507200
- Sakudoh, T., Sezutsu, H., Nakashima, T., Kobayashi, I., Fujimoto, H., Uchino, K., Banno, Y., Iwano, H., Maekawa, H., Tamura, T., Kataoka, H. and Tsuchida, K. (2007) Carotenoid silk coloration is controlled by a carotenoidbinding protein, a product of the Yellow blood gene. Proceedings of the National Academy of Sciences of the United States of America, 104, 8941-8946. doi:10.1073/pnas.0702860104
- Tsuchida, K., Jouni, E.Z., Gardetto, J., Kobayashi, Y., Tabunoki, H., Azuma, M., Sugiyama, H., Takada, N., Maekawa, H., Banno, Y., Fujii, H., Iwano, H. and Wells, A.M. (2004) Characterization of the carotenoid-binding protein of the Y-gene dominant mutants of Bombyx mori. Journal of Insect Physiology, 50, 363-372. doi:10.1016/j.jinsphys.2004.02.006
- Sakudoh, T., Tsuchida, K. and Kataoka, H. (2005) BmStart1, a novel carotenoid-binding protein isoform from Bombyx mori, is orthologous to MLN64, a mammalian cholesterol transporter. Biochemical and Biophysical Research Communications, 336, 1125-1135. doi:10.1016/j.bbrc.2005.08.241
- Tabunoki, H., Higurashi, S., Ninagi, O., Fujii, H., Banno, Y., Nozaki, M., Kitajima, M., Miura, N., Atsumi, S. and Tsuchida, K. (2004) A carotenoid-binding protein (CBP) plays a crucial role in cocoon pigmentation of silkworm (Bombyx mori) larvae. FEBS Letters, 567, 175-178. doi:10.1016/j.febslet.2004.04.067
- Hara, W., Sosnicki, S., Banno, Y., Fujimoto, H., Takada, N., Maekawa, H., Fujii, H., Wells, M.A. and Tsukhida, K. (2007) Mapping analysis of catotenoid-binding protein of Bombyx mori by restriction fragment length polymorphism. Journal of Proteome Research, 76, 149-154. doi:10.1021/pr200551e
- Niu, Y.S., Chen, Y.D., Xi, J., Sima Y.H., Duan, X.M., Liang, H.L., Gan, L.P. and Xu, S.Q. (2010) Structure and expression analysis of cbp gene in different natural colored-cocoon strains of Bombyx mori. Hereditas (Beijing), 32, 942-950. doi:10.3724/SP.J.1005.2010.00942
- Sakudoh, T., Nakashima, T., Kuroki, Y., Fujiyama, A., Kohara, Y., Honda, N., Fujimoto, H., Shimada, T., Nakagaki, M., Banno, Y. and Tsuchida, K. (2011) Diversity in copy number and structure of a silkworm morphogenetic gene as a result of domestication. Genetics, 187, 965-976. doi:10.1534/genetics.110.124982
- Niu, Y.S., Sima, Y.H. and Xu, S.Q. (2009) Correlation of CBP gene expression in silkworm with carotenoid level in larvae. Journal of Anhui Agricultural Sciences, 37, 17986-17989. http://www.cqvip.com/qk/90168X/200936/
- Thompson, J.D., Higgins, D.G. and Gibson, T.J. (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673-4680. doi:10.1093/nar/22.22.4673
- Kozak, M. (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene, 361, 13-37. doi:10.1016/j.gene.2005.06.037
- Sakudoh, T., Iizuka, T., Narukawa, J., Sezutsu, H., Kobayashi, I., Kuwazaki, S., Banno, Y., Kitamura, A., Sugiyama, H., Takada, N., Fujimoto, H., Okuda, K., Mita, K., Tamura, T., Yamamoto, K. and Tsuchida, K. (2010) A CD36-related transmembrane protein is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. Journal of Biological Chemistry, 285, 7739-7751. doi:10.1074/jbc.M109.074435
- Rubio, R.O., Suzuki, A., Mitsumasu, K., Homma, T., Niimi, T., Yamashita, O. and Yaginuma, T. (2011) Cloning of cDNAs encoding sorbitol dehydrogenase-2a and b, enzymatic characterization, and up-regulated expression of the genes in Bombyx mori diapause eggs exposed to 5˚C. Insect Biochemistry and Molecular Biology, 41, 378-387. doi:10.1016/j.ibmb.2011.02.007
- Miura, F., Kawaguchi, N., Sese, J., Toyoda, A., Hattori, M., Morishita, S. and Ito, T. (2006) A large-scale fulllength cDNA analysis to explore the budding yeast transcriptome. Proceedings of the National Academy of Sciences of the United States of America, 103, 17846-17851. doi:10.1073/pnas.0605645103
- Rodelsperger, C., Kohler, S., Schulz, H.M., Manke, T., Bauer, S. and Robinson, N.P. (2009) Short ultraconserved promoter regions delineate a class of preferentially expressed alternatively spliced transcripts. Genomics, 94, 308-316. doi:10.1016/j.ygeno.2009.07.005
- Huntington, C.J., Lam, K.C.L., Dias, C. and Majewski, J. (2009) Fine-scale variation and genetic determinants of alternative splicing across individuals. Plos Genetics, 5, e1000766. doi:10.1371/journal.pgen.1000766
- Jin, Y.F., Tian, N., Cao, J., Liang, J., Yang, Z.L. and Lv, J.N. (2007) RNA editing and alternative splicing of the insect nAChR subunit alpha6 transcript: Evolutionary conservation, divergence and regulation. BMC Evolutionary Biology, 7, 98. doi:10.1186/1471-2148-7-98
- Shao, Y.M., Dong, K. and Zhang, C.X. (2007) The nicotinic acetylcholine receptor gene family of the silkworm, Bombyx mori. BMC Genomics, 8, 324. doi:10.1186/1471-2164-8-324
- Schwartz, S., Meshorer, E. and Ast, G. (2009) Chromatin organization marks exon-intron structure. Nature Structural & Molecular Biology, 16, 990-995. doi:10.1038/nsmb.2119
- Tilgner, H., Nikolaou, C., Althammer, S., Ssmmeth, M., Beato, M., Valcarcel, J. and Guigo, R. (2009) Nucleosome positioning as a determinant of exon recognition. Nature Structural & Molecular Biology, 16, 996-1001. doi:10.1038/nsmb.1658
- Wakamatsu, A., Kimura, K., Yamamoto, J.I., Nishikawa, T., Nomura, N. Sugano, S. and Isogai, T. (2009) Identification and functional analyses of 11 769 full-length human cDNAs focused on alternative splicing. DNA Research, 16, 371-383. doi:10.1093/dnares/dsp022
- The International Silkworm Genome Consortium (2008) The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochemistry and Molecular Biology, 38, 1036-1045. doi:10.1016/j.ibmb.2008.11.004
- Adams, S.D., Erickbush, H.T., Herrera, J.R. and Lizardi, M.P. (1986) A highly reiterated family of transcribed oligo (A)-terminated, interspersed DNA elements in the genome of Bombyx mori. Journal of Molecular Biology, 187, 465-478. doi:10.1016/0022-2836(86)90327-X
ABBREVIATIONS
CBP, carotenoid binding protein;
TRAS, telomeric repeat-associated sequence;
CATS, telomeric repeat-associated sequence-like element;
AS, Alternative splicing.
NOTES
#The author contribute equally to this study.
*Corresponding author.

