Journal of Biomaterials and Nanobiotechnology
Vol.08 No.03(2017), Article ID:77632,13 pages
10.4236/jbnb.2017.83013
Microtopography Attenuates Endothelial Cell Proliferation by Regulating MicroRNAs
Dan Wang1, Mengya Liu1, Shuangying Gu1, Yue Zhou1*, Song Li1,2*
1School of Biomedical Engineering and Med-X Research Institution, Shanghai Jiao Tong University, Shanghai, China
2Department of Bioengineering and Department of Medicine, University of California, Los Angeles, CA, USA

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: March 23, 2017; Accepted: July 11, 2017; Published: July 14, 2017
ABSTRACT
Endothelial cell (EC) morphology can be regulated by the micro/nano topography in engineered vascular grafts and by hemodynamic forces in the native blood vessels. However, how EC morphology affects miRNA and thus EC functions is not well understood. In this study, we addressed this question by using human umbilical vein endothelial cells (HUVECs) cultured on microgrooves as a model. HUVECs were grown on either microgrooved (with 10 μm width/spacing and 3 μm depth) or smooth surfaces. HUVECs on microgrooved surface had elongated and bipolar morphology, while HUVECs on smooth surface showed cobble stone shape or non-polar morphology. EdU staining indicated that HUVECs with elongated morphology had lower proliferation rate compared to their counterpart cultured on smooth surface. Quantitative PCR analysis demonstrated that the expression of the specific microRNAs (miR-10a, miR-19a, miR-221) that targeted proliferation-related genes was all up-regulated. Consistently, the mRNA levels of their respective target genes, mitogen-activated protein kinase kinase kinase 7, Cyclin D1 and c-kit were significantly reduced by a fold change of 0.12 ± 0.01 (p < 0.01), 0.70 ± 0.23 (p < 0.05) and 0.76 ± 0.21 (p < 0.05). Other miRNAs such as miR-126 and miR-181a were up-regulated as well, leading to the repression of their targets vascular cell adhesion molecule-1 and prospero homeobox-1. Our results suggested that microgrooved surface may regulate microRNA levels and thus EC functions. These results provide insight into the modulation of EC functions by microtopographic cues, and will facilitate the rational design of microstructured materials for cell and tissue engineering.
Keywords:
Microgroove, PDMS, MicroRNA, Endothelial Cell

1. Introduction
Cardiovascular disease is one of the leading causes of death all around the world [1] . Vascular tissue engineering provides a valuable approach to replace diseased arteries [2] . By far, one of the main challenges in vascular tissue engineering is to enhance endothelialization on the luminal surface of vascular grafts by optimizing extracellular microenvironment [3] [4] [5] . Therefore, fine-tuning the physical and chemical properties of the scaffolds, as well as their interaction with vascular cells is pivotal to regeneration outcome and long-term effectiveness in clinical application [6] .
Many factors in the extracellular environment such as signal molecules, cell- cell adhesions and extracellular matrix affect cell behaviors. Among these factors, physical cues also play important roles in regulating cell functions. For example, micro/nano-topographic patterned surfaces of biomaterial have profound impact on cell proliferation, differentiation, migration, apoptosis and related to immunology response. For example, micro-needles regulate cell adhesion, cell lysing and cell growth [7] , and microposts accelerate the proliferation of connective tissue progenitor cells [8] . Parallel microgrooves combined with surface energy affect rat dermal fibroblasts growth [9] . Microgrooved surfaces facilitate the derivation of cardiomyocytes from stem cells [10] , affect proliferation of vascular smooth muscle cells [11] [12] , and regulate cell reprogramming and epigenetic state [13] [14] . However, whether and how microgrooved surface modulates microRNA and thus endothelial cells (EC) functions is not well understood.
It has been reported that EC functions can be regulated by microRNA [15] [16] . For example, miR-126 is essential in maintaining the proliferative capacity of EC by suppressing the Notch1 inhibitor Dlk1 and leading to EC proliferation after injury, showing a protective effect against atherosclerosis and free-radical induced apoptosis [17] [18] . Under physiological conditions in vivo, ECs are subject to laminar flow and high shear stress in the straight part of arteries, have elongated morphology, and microRNAs, including miR-10a, -19a and -126 are induced in these areas to inhibit proliferation and decrease inflammation [19] [20] .
We postulated that microgrooved surface could induce elongated EC morphology and mimick, at least in part, the effects of high shear stress on EC functions. Therefore, in this paper, we investigated whether the microtopology of the substrate may affect specific miRNA expression to regulate cell proliferation.
2. Materials and Methods
2.1. Material Preparation
We used microfabrication technique to fabricate PDMS membranes with desired surface topography. Briefly, a silicon wafer was first prepared by soft lithography. Poly(dimethyl siloxane) (PDMS, Sylgard 184, Dow Corning, Midland, MI) was spin-coated onto the patterned silicon wafers to desired thickness (~250 μm). Then the membrane was degassed under vacuum and cured at 70˚C. The membrane was detached from the wafer, cleaned by sonication, treated with oxygen plasma to enhance protein adsorption, and coated with 2% gelatin for 1 hour.
2.2. Surface Characterization
PDMS membranes were cut into 1 × 1 cm2 to fit the carrier and processed by Au sputter. The images of the samples were collected by using scanning electron microscopy (SEM) (JEOL JSM-5600, Japan). The water contact angle was determined by EasyDrop (FM40Mk2, Kruss Gmbh, Germany) and the data was analyzed by KRUSSK100 software.
2.3. Cell Culture
Human umbilical vein endothelial cells (HUVECs) were purchased from ScienCell (China). The cells were cultured with Endothelial Cell Medium (ECM, ScienCell, USA) supplemented with 5% fetal bovine serum, 1% Penicillin/ Streptomycin Solution and 1% EC growth supplement at 37˚C under 5% CO2. After reaching 80% - 90% confluency, cells were seeded on PDMS membrane for subsequent study. For consistency, cells from passages 4 - 7 were used.
2.4. Fluorescence Staining
Cells were fixed by 4% paraformaldehyde (PFA, Electron Microscopy Sciences, USA) followed by 0.5% Triton X-100 (Solarbio) to increase the permeability. Actin cytoskeleton was stained by FITC-phalloidin conjugate (AAT Bioquest) and nuclear by 4',6-diamino-2-phenylindole (DAPI, Beyotime, China). Laser scanning confocal microscope TCS SP5II (Leica) was used to acquire microscopic images.
2.5. Cell Proliferation Assay by EdU Staining
At different time points, the cells were incubated with 50 mM EdU (RiboBio Co., Ltd, China) for 2 hour at 37˚C and 5% CO2. At the end of the incubation, the cells were rinsed by phosphate buffered saline (PBS) 3 times, fixed by 4% PFA, and incubated with 2 mg/ml amino acetic acid for 5 min with slow oscillation at room temperature. Then the cells were incubated with the penetrant followed by EdU dye liquor provided in the kit for 30 min at 37˚C. Cell nuclei were stained with DAPI for cell counting. The Laser scanning microscope TCS SP5Ⅱ (Leica) was used for imaging.
2.6. Total RNA Extraction and Real-Time Polymerase Chain Reaction (PCR) Analysis
RNA was isolated from cultured cells by using the TRIzol® Plus RNA Purification Kit (No. 12183555, Thermo Fisher Scientific, USA). cDNA synthesis was performed by using the FastQuant RT kit (with gDNase) (KR106, TIANGEN, China), and SuperReal PreMix Color (SYBR Green) (FP215, TIANGEN, China) was applied for mRNA expression analysis. The primers used for real-time PCR are listed in Table 1. The annealing temperature in all the real-time PCR reaction was set at 60˚C. The gene expression level in each sample was normalized with GAPDH level respectively.
Table 1. The primers used for mRNA real-time PCR.
2.7. miRNA Preparation and Real-Time PCR Analysis
The miRcute miRNA Isolation Kit (DP501, TIANGEN, China) was used for microRNA extraction. cDNAs were synthesized by miRcute miRNA First-Strand cDNA Synthesis Kit (KR201, TIANGEN, China) and the real-time PCR was performed by miRcute miRNA qPCR Detection Kit (SYBR Green) (FP401, TIANGEN, China) according to the manufacturer’s instruction. The real-time PCR reaction was performed on Applied Biosystems 7900 HT Fast Real-Time PCR System (ABI, USA) and the data were analyzed by using SDS Software (Ver. 2.4). The expression levels of the target microRNA were normalized to U6 as the internal control. All reactions were performed in triplicates and the relative expression was calculated by comparative 2−∆∆CT method. The primers used for real-time PCR are listed in Table 2. The annealing temperature in all the real-time PCR reaction was set at 60˚C.
2.8. Statistics
Experiments were run in triplicates for each sample. Standard error was plotted as error bars in all figures. Statistically significant differences were assessed and determined by using GraphPad Prism 5. A difference with a p-value <0.05 was considered statistically significant.
3. Results
3.1. Characterization of the Microgrooved PDMS Substrates and HUVECs Morphology
PDMS is a hydrophobic material, which is not suitable for cell attachment. In order to increase its biocompatibility, 2% gelatin was used to coat the PDMS membrane surface. After coating with gelatin, the PDMS surfaces hydrophilicity were determined by water contact angle measurements. The results are shown in Figure 1 and Table 3. For PDMS membrane with smooth surface, the average contact angle decreased from 145.567 to 71.267 degree, and for the microgroove pattern, the contact angle decreased from 125.967 to 72.433 degree. It is clear that gelatin coating significantly increased the hydrophilicity of the PDMS membrane. Since the water droplet was much larger than the size of the microgrooves, no significant difference was observed between the smooth surface and microgrooved surface.
Figure 1. Water contact angle. The water contact angle of both smooth and grooved surface of PDMS membrane before and after gelatin coating following the process of plasma treatment.
Table 2. The primers used for microRNA real-time PCR.
Table 3. Contact angle of both smooth and grooved surface of PDMS membrane.
To confirm that coating with gelatin did not change the membrane surface topography, the PDMS surfaces were also characterized by scanning electron microscopy (SEM). As shown in Figure 2(a), the micropatterned PDMS surface consists of 10-μm wide and 3-μm deep microgrooves. Therefore, the coating did not change the desired surface topography of the grooved PDMS membrane.
HUVECs were seeded on both surfaces for 3 days (Figure 2(b)) and stained with phalloidin and DAPI before confocal imaging. On the smooth PDMS membrane, the cells spread out and had thick bundles of F-actin or stress fibers. On the contrary, the cells grew on microgrooved PDMS membrane demonstrated an elongated shape, as the cytoskeleton arranged align in the direction of the grooves.
3.2. Microgroove Reduced the Proliferation of HUVECs
Next, we performed EdU staining to quantify cell proliferation (Figure 3). For cells cultured on smooth surface, proliferation rate was similar at the second and

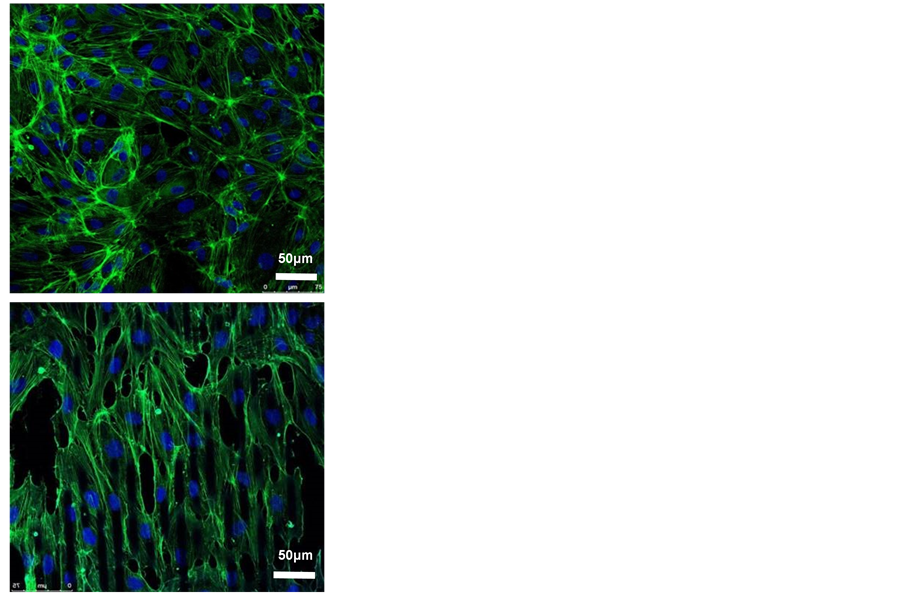
Figure 2. Effects of micropattern on the morphology of HUVECs. (a) SEM micrograph of PDMS membranes with smooth or microgroove surface. The parallel microgrooved were 10 μm in width and 3 μm in depth, magnification = 100; (b) HUVECs stained by phalloidin (green) and DAPI (blue), magnification = 40, scale bar = 50 μm.
third day after seeding. However, for cells cultured on microgrooved surface, the proliferation rate was significantly lower on day 2 (p < 0.01) and day 3 (p < 0.05) than the cells seeded on smooth surface.
3.3. Microgroove Substrate Regulated the Expression of miRNAs and Their Target Genes Related to EC Proliferation
We further investigated the microRNAs that had been reported to regulate EC proliferation. As shown in Figure 4(a), miR-10a, miR-19a and miR-221 were up-regulated in HUVECs cultured on microgrooved surface by a fold change of 7.67 ± 1.11 (p < 0.01), 9.76 ± 3.83 (p < 0.05) and 1.92 ± 0.03 (p < 0.01), respectively. Based on previous findings, miR-10a attenuates the expression of mitogen-activated kinase kinase kinase 7 (MAP3K7; TAK1) by binding to the 3’ UTR of its gene and then affects IκB/NF-κB pathway [21] . MiR-19a can significantly decrease the protein level of cyclin D1 (CCND1), leading to an arrest of cell cycle at G1/S transition [22] . Unlike the role in cancer cells, miR-221 reduce c-kit expression in EC and thus inhibit cell proliferation and vessel growth [23] . All the above microRNAs, with their up-regulation attenuating the expressions of their target genes, may lead to the inhibition of EC proliferation.
Therefore, to confirm this, the target genes of miR-10a, miR-19a and miR-221 were specifically selected and real-time PCR was used to examine the gene ex-

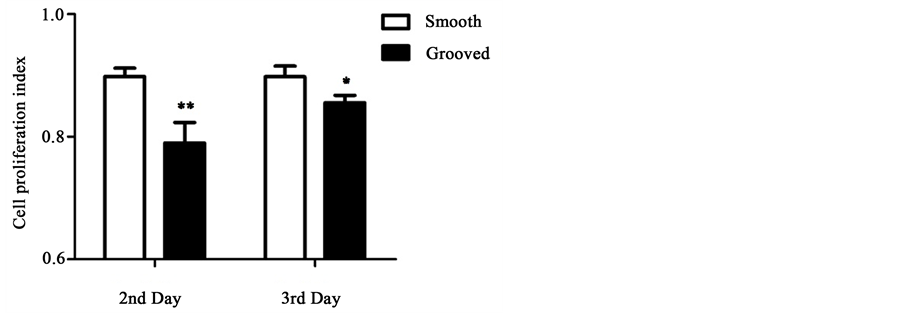
Figure 3. EdU staining of cells cultured on smooth or microgrooved PDMS membranes. (a) HUVECs were cultured for 2-3 days, and stained by EdU (green) for proliferated cells and by DAPI (blue) for all cell nuclei; (b) The proliferation index of cells cultured on smooth or microgrooved surface. The data were shown as the means ± SE. n = 3, *p < 0.05, **p < 0.01.
pression level at day 3. Previous work has shown that MAP3K7 is a key regulator of IκBα, which is a core member of IκB family. A sustained binding of IκB and NF-κB will cause a lower growth rate of multiple types of cells including ECs [24] . Furthermore, over-expressed CCND1 binds to and activates CDK4 causing hyperphosphorylation of RB1, which promotes cell cycle G1/S transition [25] . And c-kit protein is a member of trans-membrane receptor tyrosine kinase (RTK) family that is activated by stem cell factor to promote cell activity including proliferation [26] . As shown in Figure 4(b), cell cycle-related genes MAP3K7,

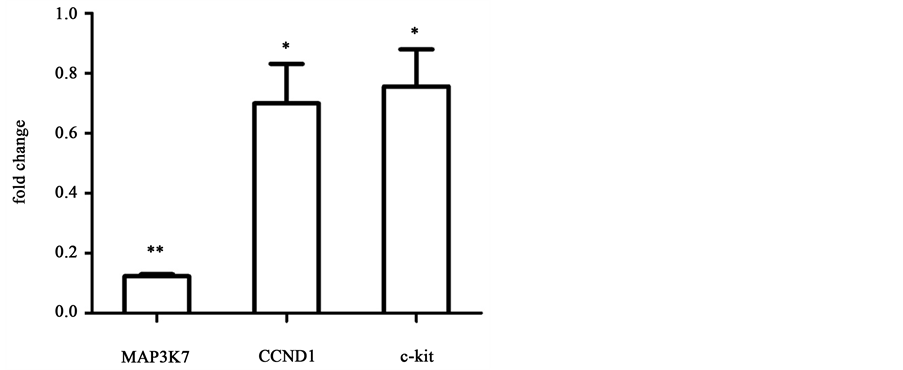
Figure 4. Real-time PCR analysis of growth-related miRNAs and target genes for cells cultured on microgrooved or smooth PDMS membrane. Results were normalized to smooth surface as control, n = 3, *p < 0.05, **p < 0.01.
CCND1 and c-kit, predicted as the target genes of miR-10a, miR19a and miR- 221 respectively, were all down-regulated on microgrooved surfaces compared to that on smooth surfaces and the fold change values are 0.12 ± 0.01 (p < 0.01), 0.70 ± 0.23 (p < 0.05) and 0.76 ± 0.21 (p < 0.05), respectively, consistent with the finding on cell proliferation.
3.4. Microgroove Substrate Regulated the Expression of miRNAs and Their Target Genes Related to Other EC Functions
We also investigated the microRNAs that had been reported to participate in the regulation of EC inflammation and differentiation. As shown in Figure 5(a), miR-126 and miR-181a were both up-regulated in the HUVECs cultured on microgrooved surface by a fold change of 5.46 ± 0.86 (p < 0.01) and 2.41 ± 0.82 (p < 0.05). Based on previous findings, miR-126 is one of the most abundant microRNAs in ECs and involves in diverge biological processes, such as anti-in-


Figure 5. Real-time PCR analysis of other miRNAs and target genes for cells cultured on microgrooved or smooth PDMS membrane. Results were normalized to smooth surface as control, n = 3, *p < 0.05, **p < 0.01.
flammation, angiogenesis and vascular remodeling under shear stress [27] [28] [29] . By looking at the inflammatory biomarkers, we found that vascular cell adhesion molecule 1 (VCAM-1), a target of miR-126, was reduced and the fold change is 0.71 ± 0.12 (p < 0.05), as shown in Figure 5(b). In addition, the expression of Prospero Homeobox 1(PROX1), a target gene of miR-181a, was also reduced by a fold change of 0.57 ± 0.03 (p < 0.01). PROX1 expresses during vascular development and neo-lymphangiogenesis [30] . miR-181a binds to its 3' UTR resulted in reduced levels of PROX1 and then reprogramming of lymphatic ECs toward a blood vascular phenotype.
4. Discussion and Conclusions
It is known that atherosclerotic plaques occur in bifurcation of vessels or curved arteries, where ECs are exposed to disturbed flow, featuring low speed and oscillatory shear stress. In addition, vascular ECs sense and respond to blood flow, ultimately, change gene expression [31] [32] . The shape of ECs exposed to laminar shear stress is elongated and unidirectional along with the flow direction. From this point of view, the physical effect of microgrooved surface is similar to that of laminar shear stress, at least in some ways. Indeed, cell proliferation on microgrooves or in response to shear stress decreases. Therefore, tissue engineered blood vessel with aligned micro or nano patterns might mimic the effect of higher shear stress on ECs, and ECs on aligned matrix, upon implantation into low-speed blood flow area, may still behave as they were under high shear stress blood flow, and are thus inhibitory for atherosclerotic lesion formation.
Accumulative evidence indicates that so called “mechanical microRNAs” are responsible for the flow-dependent control of endothelial function [19] . Anti- atherogenic mechanical microRNAs such as miR-10a and 19a are either up-re- gulated by high shear stress or are down-regulated by low shear stress in vivo [33] . miR-10a inhibits NF-κB through stabilizing IκB/NF-κB complex by targeting MAP3K7 and βTRC that are the key factors for IκB degradation, and thus a variety of cell behaviors are affected, including proliferation. Correspondingly, in our study, after 3 day of seeding, miR-10a was upregulated and the proliferation of HUVECs was attenuated.
Previous studies on HUVEC in vitro culture system determined flow-sensi- tive miRNAs; after 12 h of shear stress (12 dyn/cm2) treatment, miR-19a, which targets CCND1, was identified as an upregulated miRNA. Consistently, after 3 days of seeding, just like miR-10a, miR-19a was upregulated and the proliferation of HUVECs was inhibited in our study.
miR-221 is a multiple target microRNA. c-kit, p27, p57 are all targets of miR- 221/222 cluster. p27 and p57 are both tumor suppressor genes. High expression of miR-221 in cancer cells promotes cell proliferation by targeting the cell cycle inhibitor p27 [34] . However, c-kit was reported to be strongly expressed in ECs, whereas both p27 and p57 expressed weakly. Therefore, high expression of miR-221 in EC showed an opposite effect to cancer cells.
As a mechanical microRNA, miR-126 has been implicated in either pro- or anti-atherogenic events, and the role is determined by lots of variables which influence cell sensitivity to the laminar flow. In our study, we detected up-regula- tion of miR-126 induced by microgrooved surface and reduced expression of VCAM-1, which imply a possible anti-inflammation effect of EC on microgrooved surface.
The proliferation of HUVECs can be regulated by numerous factors; in our study, microgrooved PDMS membrane exert the proliferation inhibitory effect on HUVEC by up-regulation of a variety of microRNAs, the high level of miR- 10a, -19a, -221 leading to down-regulation of MAP3K7, CCND1 and c-kit, which were key factors during the proliferation process. The method of modulating material surface morphology to interfere cell behavior, or specifically, using scaffold with microgrooved surface to affect EC proliferation, might be used in engineer surface in blood vessel tissue engineering.
Acknowledgements
This work was supported in part by the Ministry of Science and Technology (2016YFC1100202), the Science and Technology Commission of Shanghai Municipality (16140901900). SL was also supported by grants from the National Institute of Health (HL117213 and HL121450).
Cite this paper
Wang, D., Liu, M.Y., Gu, S.Y., Zhou, Y. and Li, S. (2017) Microtopography Attenuates Endothelial Cell Proliferation by Regulating MicroRNAs. Journal of Biomaterials and Nanobiotechnology, 8, 189-201. https://doi.org/10.4236/jbnb.2017.83013
References
- 1. Alexopoulos, N. and Raggi, P. (2009) Calcification in Atherosclerosis. Nature Reviews Cardiology, 6, 681-688.
https://doi.org/10.1038/nrcardio.2009.165 - 2. Wessely, R. (2010) New Drug-Eluting Stent Concepts. Nature Reviews Cardiology, 7, 194-203.
https://doi.org/10.1038/nrcardio.2010.14 - 3. Engler, A.J., Sen, S., Sweeney, H.L. and Discher, D.E. (2006) Matrix Elasticity Directs Stem Cell Lineage Specification. Cell, 126, 677-689.
https://doi.org/10.1016/j.cell.2006.06.044 - 4. Murphy, W.L., McDevitt, T.C. and Engler, A.J. (2014) Materials as Stem Cell Regulators. Nature Materials, 13, 547-557.
https://doi.org/10.1038/nmat3937 - 5. Dalby, M.J., Gadegaard, N. and Oreffo, R.O. (2014) Harnessing Nanotopography and Integrin-Matrix Interactions to Influence Stem Cell Fate. Nature Materials, 13, 558-569.
https://doi.org/10.1038/nmat3980 - 6. Liang, C., Hu, Y., Wang, H., Xia, D., Li, Q., Zhang, J., et al. (2016) Biomimetic Cardiovascular Stents for in Vivo Re-Endothelialization. Biomaterials, 103, 170-182.
https://doi.org/10.1016/j.biomaterials.2016.06.042 - 7. Tan, J.L., Tien, J., Pirone, D.M., Gray, D.S., Bhadriraju, K. and Chen, C.S. (2003) Cells Lying on a Bed of Microneedles: An Approach to Isolate Mechanical Force. Proceedings of the National Academy of Sciences of the United States of America, 100, 1484-1489.
https://doi.org/10.1073/pnas.0235407100 - 8. Kim, E.J., Boehm, C.A., Mata, A., Fleischman, A.J., Muschler, G.F. and Roy, S. (2010) Post Microtextures Accelerate Cell Proliferation and Osteogenesis. Acta Biomaterialia, 6, 160-169.
https://doi.org/10.1016/j.actbio.2009.06.016 - 9. den Braber, E.T., de Ruijter, J.E., Smits, H.T., Ginsel, L.A., von Recum, A.F. and Jansen, J.A. (1995) Effect of Parallel Surface Microgrooves and Surface Energy on Cell Growth. Journal of Biomedical Materials Research, 29, 511-518.
https://doi.org/10.1002/jbm.820290411 - 10. Morez, C., Noseda, M., Paiva, M.A., Belian, E., Schneider, M.D. and Stevens, M.M. (2015) Enhanced Efficiency of Genetic Programming toward Cardiomyocyte Creation through Topographical Cues. Biomaterials, 70, 94-104.
https://doi.org/10.1016/j.biomaterials.2015.07.063 - 11. Thakar, R.G., Cheng, Q., Patel, S., Chu, J., Nasir, M., Liepmann, D., et al. (2009) Cell-Shape Regulation of Smooth Muscle Cell Proliferation. Biophysical Journal, 96, 3423-3432.
https://doi.org/10.1016/j.bpj.2008.11.074 - 12. Vernon, R.B., Gooden, M.D., Lara, S.L. and Wight, T.N. (2005) Microgrooved Fibrillar Collagen Membranes as Scaffolds for Cell Support and Alignment. Biomaterials, 26, 3131-3140.
https://doi.org/10.1016/j.biomaterials.2004.08.011 - 13. Sia, J., Yu, P., Srivastava, D. and Li, S. (2006) Effect of Biophysical Cues on Reprogramming to Cardiomyocytes. Biomaterials, 103, 1-11.
https://doi.org/10.1016/j.biomaterials.2016.06.034 - 14. Downing, T.L., Soto, J., Morez, C., Houssin, T., Fritz, A., Yuan, F., et al. (2013) Biophysical Regulation of Epigenetic State and Cell Reprogramming. Nature Materials, 12, 1154-1162.
https://doi.org/10.1038/nmat3777 - 15. Zhang, C. (2010) MicroRNAs in Vascular Biology and Vascular Disease. Journal of Cardiovascular Translational Research, 3, 235-240.
https://doi.org/10.1007/s12265-010-9164-z - 16. Peng, Y.J., Ren, Z., He, N.Y., Li, Q., Zhao, X.S., et al. (2016) Functional Regulatory Roles of MicroRNAs in Atherosclerosis. Clinica Chimica Acta, 460, 164-171.
https://doi.org/10.1016/j.cca.2016.06.044 - 17. Schober, A., Nazari-Jahantigh, M., Wei, Y., Bidzhekov, K., Gremse, F., Grommes, J., et al. (2014) MicroRNA-126-5p Promotes Endothelial Proliferation and Limits Atherosclerosis by Suppressing Dlk1. Nature Medicine, 20, 368-376.
https://doi.org/10.1038/nm.3487 - 18. Sui, X.Q., Xu, Z.M., Xie, M.B. and Pei, D.A. (2014) Resveratrol Inhibits Hydrogen Peroxide-Induced Apoptosis in Endothelial Cells via the Activation of PI3K/Akt by MiR-126. Journal of Atherosclerosis and Thrombosis, 21, 108-118.
https://doi.org/10.5551/jat.19257 - 19. Kumar, S., Kim, C.W., Simmons, R.D. and Jo, H. (2014) Role of Flow-Sensitive MicroRNAs in Endothelial Dysfunction and Atherosclerosis: Mechanosensitive Athero-MiRs. Arteriosclerosis, Thrombosis, and Vascular Biology, 34, 2206-2216.
https://doi.org/10.1161/ATVBAHA.114.303425 - 20. Araldi, E. and Suarez, Y. (2016) MicroRNAs as Regulators of Endothelial Cell Functions in Cardiometabolic Diseases. Biochimica et Biophysica Acta, 1861, 2094-2103.
https://doi.org/10.1016/j.bbalip.2016.01.013 - 21. Fang, Y., Shi, C., Manduchi, E., Civelek, M. and Davies, P.F. (2010) MicroRNA-10a Regulation of Proinflammatory Phenotype in Athero-Susceptible Endothelium in Vivo and in Vitro. Proceedings of the National Academy of Sciences of the United States of America, 107, 13450-13455.
https://doi.org/10.1073/pnas.1002120107 - 22. Qin, X., Wang, X., Wang, Y., Tang, Z., Cui, Q., Xi, J., et al. (2010) MicroRNA-19a Mediates the Suppressive Effect of Laminar Flow on Cyclin D1 Expression in Human Umbilical Vein Endothelial Cells. Proceedings of the National Academy of Sciences of the United States of America, 107, 3240-3244.
https://doi.org/10.1073/pnas.0914882107 - 23. Liu, X., Cheng, Y., Yang, J., Xu, L. and Zhang, C. (2012) Cell-Specific Effects of MiR-221/222 in Vessels: Molecular Mechanism and Therapeutic Application. Journal of Molecular and Cellular Cardiology, 52, 245-255.
https://doi.org/10.1016/j.yjmcc.2011.11.008 - 24. Tobon-Arroyave, S.I., Hurtado-Garcia, P., Garcia-Quintero, O.D., Isaza-Guzman, D.M. and Florez-Moreno, G.A. (2015) Immunoexpression of NF-KB and Their Inhibitory Subunits IkBalpha and IkBbeta in Giant Cell Lesions of the Jaws: Implications for Their Clinical Behavior. Journal of Oral Pathology & Medicine: Official Publication of the International Association of Oral Pathologists and the American Academy of Oral Pathology, 44, 752-760.
https://doi.org/10.1111/jop.12289 - 25. Desidero, D.T., Fioravanti, A., Orlandi, P., Canu, B., Giannini, R., Borrelli, N., et al. (2013) Antiproliferative and Proapoptotic Activity of Sunitinib on Endothelial and Anaplastic Thyroid Cancer Cells via Inhibition of Akt and ERK1/2 Phosphorylation and by Down-Regulation of Cyclin-D1. The Journal of Clinical Endocrinology and Metabolism, 98, E1465-1473.
https://doi.org/10.1210/jc.2013-1364 - 26. Liu, X., Cheng, Y., Zhang, S., Lin, Y., Yang, J. and Zhang, C.A. (2009) Necessary Role of MiR-221 and MiR-222 in Vascular Smooth Muscle Cell Proliferation and Neointimal Hyperplasia. Circulation Research, 104, 476-487.
https://doi.org/10.1161/CIRCRESAHA.108.185363 - 27. Tang, S.T., Wang, F., Shao, M., Wang, Y. and Zhu, H.Q. (2017) MicroRNA-126 Suppresses Inflammation in Endothelial Cells under Hyperglycemic Condition by Targeting HMGB1. Vascular Pharmacology, 88, 48-55.
https://doi.org/10.1016/j.vph.2016.12.002 - 28. Esser, J.S., Saretzki, E., Pankratz, F., Engert, B., Grundmann, S., Bode, C., et al. (2017) Bone Morphogenetic Protein 4 Regulates MicroRNAs MiR-494 and MiR-126-5p in Control of Endothelial Cell Function in Angiogenesis. Thrombosis and Haemostasis, 117, 734.
https://doi.org/10.1160/TH16-08-0643 - 29. Mondadori, S.A., Metzinger, L., Haddad, O., M'Baya-Moutoula, E., Taibi, F., Charnaux, N., et al. (2015) MiR-126 Is Involved in Vascular Remodeling under Laminar Shear Stress. BioMed Research International, 2015, Article ID: 497280.
https://doi.org/10.1155/2015/497280 - 30. Kazenwadel, J., Michael, M.Z. and Harvey, N.L. (2010) Prox1 Expression is Negatively Regulated by MiR-181 in Endothelial Cells. Blood, 16, 2395-2401.
https://doi.org/10.1182/blood-2009-12-256297 - 31. Chiu, J.J. and Chien, S. (2011) Effects of Disturbed Flow on Vascular Endothelium: Pathophysiological Basis and Clinical Perspectives. Physiological Reviews, 91, 327-387.
https://doi.org/10.1152/physrev.00047.2009 - 32. Ando, J. and Yamamoto, K. (2011) Effects of Shear Stress and Stretch on Endothelial Function. Antioxidants & Redox Signaling, 15, 1389-1403.
https://doi.org/10.1089/ars.2010.3361 - 33. Neth, P., Nazari-Jahantigh, M., Schober, A. and Weber, C. (2013) MicroRNAs in Flow-Dependent Vascular Remodelling. Cardiovascular Research, 99, 294-303.
https://doi.org/10.1093/cvr/cvt096 - 34. Nassirpour, R., Mehta, P.P., Baxi, S.M. and Yin, M.J. (2013) MiR-221 Promotes Tumorigenesis in Human Triple Negative Breast Cancer Cells. PloS One, 8, e62170.
https://doi.org/10.1371/journal.pone.0062170



