Agricultural Sciences
Vol.3 No.8(2012), Article ID:25453,8 pages DOI:10.4236/as.2012.38118
RT-PCR and CP gene based molecular characterization of a cucumber mosaic cucumovirus from Aligarh, U.P., India
![]()
1SNPG College, VBS Purvanchal University, Jaunpur, India; *Corresponding Author: s.ali@cgiar.org
2Central Potato Research Station (CPRS), Indian Council of Agricultural Research (ICAR), Meghalaya, India
3Plant Virology Laboratory, Department of Botany, A.M.U., Aligarh, India
Received 2 August 2012; revised 9 September 2012; accepted 19 October 2012
Keywords: Salvia; Mosaic; Non-persistent; DAC-ELISA; RT-PCR; Cucumovirus
ABSTRACT
A virus disease of garden sage (Salvia splendens Ker-Gawl.) was observed and characterized showing symptoms of severe mosaic, mottling and distortion of leaves being remain shortened and growth retarded. The virus was transmitted to the healthy plants of Salvia spp. as well as many other hosts by mechanical inoculation, Myzus persicae Sulzer and Aphis gossypii Glover transmit the virus in non-persistent manner. Purified sample in EM showed spherical particles c.28 nm in diameter. DAC-ELISA [1] was performed with crude sap, specific polyclonal antiserum (PVAS 242a, ATCC, USA) and alkaline phosphatase-linked secondary antibodies (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH or DSMZ, Germany). The mean absorbance at 405 nm for negative and positive controls were 0.061 ± 0.008 and 0.349 ± 0.003 respectively, while infected samples were recorded four-times more than the value of negative controls with values that ranged between 0.289 ± 0.005 and 0.325 ± 0.003. RT-PCR was performed using total RNA as templates and CMV Coat Protein (CP) gene specific reverse and forward primers, gel was electrophoresed on 1% agarose, an amplification of expected size 650 bp fragment was obtained only in the infected sample which proved that the present virus is a strain of CMV, the type member of the genus cucumovirus belonging to the family Bromoviridae.
1. INTRODUCTION
Salvia is a large genus of aromatic and ornamental herbs and shrubs distributed in temperate and tropical regions. About 24 species are known to occur in India. Among them Salvia splendens Ker-Gawl; S. officinalis L., S. farinacea Benth. and S. patens are widely grown in Indian gardens. The whole plant or different parts of the plant are used in medicine. The oil extracted from Salvia spp. i.e. sage oil is used as antibacterial, antiviral, antispasmodic and also violent epileptiform convulsant.
During the survey of viral disease on ornamental plants in literature it was found that the virus reported on Salvia sclarea was CMV [2] from Italy. Symptoms were also observed during disease evaluation on Salvia uliginosa i.e. Bog Salvia [3]. An isolate of Potato virus-Y tuber necrotic strain was also reported on Salvia sclarea [4] and Datura Stramonium Linn. as a natural host.
Cucumber Mosaic Virus (CMV) of family Bromoviridae is one of the most important widespread viruses in the world infecting the largest number (approximately 1000) of plant species [5]. The genome of the virus consists of positive-sense single stranded three RNAs (RNA 1, RNA 2 and RNA 3) and a sub-genomic RNA (RNA 4) which is encoded by the 3’-half of RNA 3 [6] and which is involved in encapsidation [7].
A number of CMV strains reported from all over the world have been placed in two subgroups I and II on the basis of serology [8-10], nucleic acid hybridization [11], gene sequences [12,13] and RFLP [14,15]. CMV subgroup I has recently been subdivided into IA and IB on the basis of gene sequence available for CMV strains and phylogenetic analysis [16-18]. Further asian strains of CMV have been placed in subgroup IB [16].
2. MATERIALS AND METHODS
Transmission Studies
Mechanical transmission: Naturally infected leaves were used for preparing inoculum with 0.1 M phosphate buffer pH 7.0. Pure culture was maintained by obtaining single lesion from Chenopodium amaranticolor Coste & Reyn and inoculating it on to Nicotiana glutinosa L. The virus was further multiplied on the hosts such as N. tabacum cv. Xanthi and N. tabacum cv. Harrison spl. For hosts range studies different hosts belonging to different families were grown in an insect proof glass house and screened against the virus obtained from pure culture.
Transmission by aphids: Healthy colony of aphids namely Myzus persicae Sulzer and A. gossypii Glover were raised in cages and the healthy nymphs of these aphids were used for transmission. The aphids were given a pre-acquisition starvation of 2 h, acquisition access 2 minutes, inoculation access 2 h using at least 10 aphids/plant and 10 recipient plants for each aphid species.
Purification: The present virus was purified [19] with slight alteration in pH of buffer and molarity. About 200 grams of infected leaves of N. tabacum cv. Samsun inoculated 10 days earlier were harvested and homogenized in 400 ml of phosphate buffer pH 7.0 of the same molarity containing 0.1% of thioglycolic acid and 0.1% sodium sulphite. Clarification of the extract and virus precipitation with 6% PEG was done [20]. The purified virus obtained from method given above was diluted with the phosphate buffer having the same molarity. After making dilution UV spectra of the virus characteristic of nucleoprotein was obtained with DU-6 spectrophotometer.
Electron microscopy: The electron microscopic studies were carried out by using clarified virus concentrate prepared [21]. Antisera of different plant viruses viz. Potato Leaf Roll Virus (PLRV), Cowpea mosaic virus (CPMV), Soybean stunt strain (CMV-SS), Pea strain (CMV-P) and MMV strain were used in IEM studies. All the antisera were diluted 200 folds with 0.88% NaCl solution before use. Trapping, decoration and mounting of trapped virions was done according to Garg and Khurana [22,23].
Serology: The identification of the CMV subgroup specific antibodies along with alkaline phosphataselinked antibodies was used for ELISA [1].
Total RNA extraction and RT-PCR: The total RNA isolated from infected leaves of Salvia splendens and Nicotina glutinosa by using RNAqueous (Ambion, USA). Viral RNA was isolated by disrupting the particles by 1% SDS followed by extraction with Phenol-chloroform method [24]. Reverse Transcription Polymerase Chain Reaction (RT-PCR) was performed to amplify the CP gene of present isolate with some flanking regions using total RNA (approximately 5 mg) and viral RNA (approximately 2 - 3 mg) both as template and CP gene specific forward and reverse primers [25]. First strand cDNA synthesis was performed using total RNA, 50 pM reverse primers and 200 µ Moloney Murine Leukemia virus reverse transcriptase enzyme in 20 µL reaction mixture, incubated at 42˚C for 90 minutes. PCR was carried out in 50 µL reaction using 5 µL cDNA as a template, 3 µL Taq DNA polymerase, 25 pM forward and reverse primers each, 10mM dNTPs of each in GeneAmp PCR 9700 system thermal cycler (Applied Biosystem, USA). The procedure is as follows: initial denaturation at 94˚C for 3 minutes followed by 30 cycles of 94˚C for 15 S, 54˚C for 30 S and 72˚C for 30 S with a final extension at 72˚C for 5 minutes.
3. RESULTS AND DISCUSSION
Transmission Studies
Mechanical inoculation made it possible to infect several hosts including Amaranthus gracilis, A. retroflexus, Antirrhinum majus, Apium graveolens, Arachis hypogea, Beta vulgaris, Brassica campestris, B. juncea, Capsicum annuum, Cucumis sativus, C. melo, Cucurbita pepo, Citrullus vulgaris, Physalis peruviana, Datura metel, Daucus carota, Dianthus barbatus, Gomphrena globosa, Lycopersicon esculentum, Pisum sativum, Raphamus sativus, Solanum melongena, S. nigrum and Spinacea oleracea showed symptoms or were latently infected (Table 1).
In aphid transmission tests Myzus persicae and Aphis gossypii transmitted the virus to 8/10, 5/10 plants respectively. The symptoms produced were identical to those induced by sap inoculation but Bemisia tabaci was failed to transmit the disease.
Mechanical inoculation from naturally infected Salvia plants transmitted the virus to C. amranticolor and C. album produced chlorotic local lesions (Figure 1) followed by systemic infection (Figure 2) while on C. quinova and C. murale the virus produced necrotic lesions (Figure 3). Symptoms on other species were systemic, mild to severe mosaic, and leaf deformation except C. pepo which produced only mild yellowing on the emerging leaves (Figure 4).
N. glutinosa and C.amaranticolor produced systemic symptoms of mosaic mottling and downward curling (Figures 5(a) and (b)) while Brassica oleracea showed upward curling of the leaves (Figure 5(c)). Systemic symptoms appeared on N. rustica in the form of green mosaic and at advance stage of infection plants showed deformation of leaves, reduced leaf lamina and growth retardation. Inoculated Capsicum annuum leaves showed necrotic lesions after 3 - 5 days of inoculation (Figure 6) Cucumis melo and Citrullus vulgaris produced discrete local lesions when the virus is inoculated on cotyledons which on later stage produced mosaic on young emerging leaves (Figure 7).
The following species which failed to develop symptoms after inoculation and virus was not recovered on
Table 1. Host range of different virus isolates.
Abbreviations: LL = Local Lesion; CL = Leaf Curling; CLL = Chlorotic Local Lesion; M = Mosaic; Mo = Mottling; MM = Mild Mosaic; VC = Vein Clearing; VB = Vein Banding; LD = Leaf Deformation; BL = Blister Like; LP = Line Pattern; CS = Chlorotic spots; DLL = Discrete Local Lesions.

Figure 1. Chlorotic local lesions.

Figure 2. Coalesces on C. amranticolor.

Figure 3. Necrotic lesions on C. quinova.

Figure 4. Mild yellowing on emerging leaf.
 (a)
(a) (b)
(b) (c)
(c)
Figure 5. (a) Mottling on N. glutinosa; (b) Down curling of Chenopodium leaf; (c) Upward curling of B. oleracea.

Figure 6. Necrotic lesions on on C. annuum.

Figure 7. Mosaic on C. pepo leaf.
back inoculation include Ageratum maxicanum, Chrysanthemum indicum, Cosmos bipinnata, Dahlia pinnata, Benincasa cerifera, Curcurbita maxima, Lagenaria siceraria, Trichosanthus anguina, Alium cepa, Althea rosea, Lathyrus odoratus, Dolicos lablab, Phlox and Carolina.
Particle Morphology: The Purified preparation of the virus obtained from the infected leaves of Nicotiana glutinosa L. showed isometric virus particles of size c. 28 nm that is typical of CMV (Figure 8). Trapping, decoration and counting of the trapped virions was done according to Garg and Khurana [22] (Figure 9).
ELISA: A high contrast of colour (yellowish green) showing decrease in intensity with increase in dilutions developed by the CMV infected plants. The mean absorbance values at 405 nm.
PCR: The result revealed the amplification of ~650 bp size band from the Salvia isolate, but not from asymptomatic when subjected to electrophoresis. The strong evidence was obtained when the infected samples was subjected to southern hybridization using α32P labeled DNA probe prepared from a CMV CP clone (EU140547). However, no such hybridization signal were found with healthy samples. The amplified product was gel purified (Au-Prep Sigma gel extraction kit) and cloned in pGEM-T easy vector system (Promega, USA). Three positive clones were sequenced and the data were evaluated for a consensus sequence and submitted to GeneBank database (Accession number EU 600215).
Basic Local Alignment Tool (BLAST) analysis of the CP gene of the Salvia isolate revealed the maximum (97%) nucleotide (nt) identity with CMV isolate of sub group IB (EF 178298 and DQ 910858), compared with medium identity (89% - 88%) with isolate of CMV sub group IA (D 01538 and D 12499) and minimum identity (70% - 67%) with sub group II (M 21464 and L 15336) CMV sequences (Table 2). Multiple nucleotide and de-
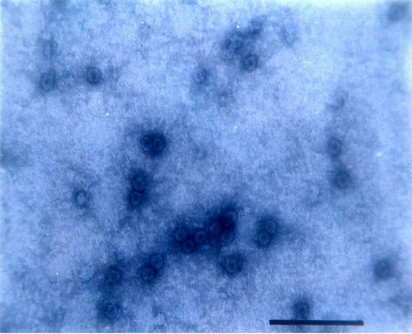
Figure 8. Isometric particle size of c. 28 nm.
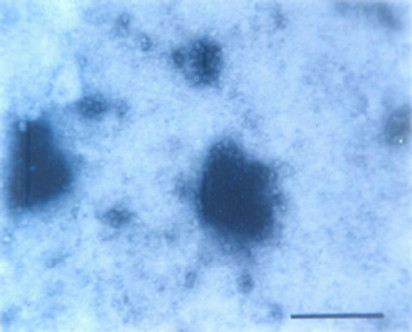
Figure 9. Trapped isometric particle of CMV.
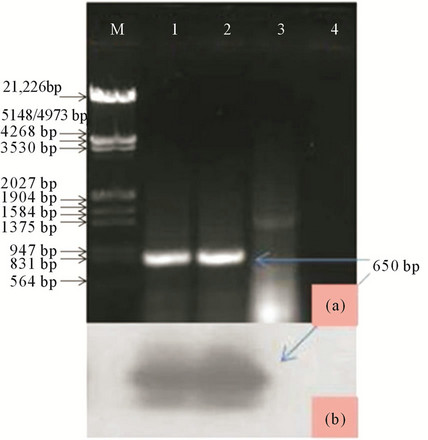
Figure 10. (a) Lane M: Lamda DNA digested with EcoRI and HindIII as DNA marker; Lane 1: CMV positive control; Lanes 2 and 3: Salvia infected and apparently healthy samples; Lane 4: RT-PCR without template (b) Southern hybridization of the same gel using specific probe.
duced amino acid (aa) alignment with the available sequence were performed using CLUSTAL-W program v1.82 [25] and aligned files were bootstrapped 100 times generating a neighbour-joining a phylogenetic tree using tree explorer. The CMV isolate also showed close phylogenetic relation ship with an Indian strain of CMV of sub group IB, where as more distant relation ships were observed for subgroup I and II, when analysed by Mega 4 [26]. These biological, serological and molecular studies confirmed the isolate under study is CMV member of sub group IB.
4. DISCUSSION
Based on DAC-ELISA, RT-PCR and phylogenetic analysis of CP gene results indicate that the virus concerned was an isolate of CMV that was serologically related to CMV-K8 and CMVSS but not with the other antisera. Two common species e.g. M. persicae and A. gossypii are able to transmit the disease in non-persistent manner. Although CMV previously has been reported to infect the genus Salvia naturally, however, such reports are rare. Therefore, the use of host plants and symptomatology to define strain is not practical. The symptom expression can vary according to temperature, age of indicator plants and source of inoculums. CMV concentration can vary between plants and between leaves of the same plants [28] and a single amino acid in coat protein can also alter the development and diversity of symptoms [29,30]. Several lines of evidence show that the virus concentration in C. murale, D. metel and also in experimentally infected C. quinova is low. The virus detected in the crude sap of these plants by biophysical means, serology and by external morphology.
The virus produced local lesions on C. amaranticolor and C. quinova, Earlier workers [2,27] also reported that their isolate from Salvia is a strain of CMV. However, the Aligarh isolate is different from the isolate reported by these earlier workers in their biophysical properties. The isolate of CMV reported by [3] from A. majus, S. splendens and S. uliginosa was compared with CMV of Aligarh isolate from S. officinalis but found to be different from CMV of above workers in their host reactions and also some variations in their bio-physical properties. However, the serological studies based on immuno-electron microscopy further revealed that the virus isolated
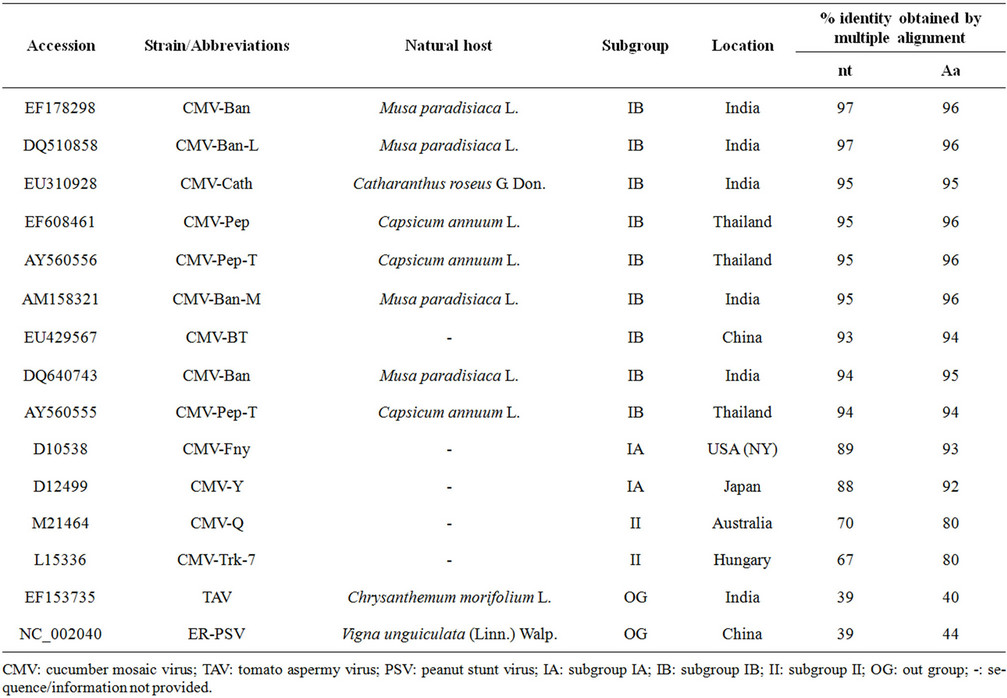
Table 2. Percentage nucleotide (nt) and amino acid (aa) identity of CMV Salvia isolate (accession No. EU 600215) with the selected CMV strains of subgroup IA, IB and II with other member of cucumovirus.
from Salvia at Aligarh has sero-affinity with CMV-SS and CMV-K8 but not with CMV-P and CMV-M, which shows that the isolate from Salvia is a strain of Cucumber mosaic virus of cucumo virus group.
REFERENCES
- Verma, N., Singh, L., Singh, A.K., Kulshreshtha, S., Raikhy, G., Hallan, V., Ram, R. and Zaidi, A.A. (2005) Ornithogalum: A new host of Cucumber Mosaic Virus (CMV) from India. Plant Pathology, 54, 256. doi:10.1111/j.1365-3059.2005.01128.x
- Pisi, A. and Vicchi, V. (1989) Infection by Cucumber Mosaic Virus (CMV) on Salvia sclarea in Italy. Informatore fitopathologico, 39, 49-51.
- Holcomb, G.E., Buras, H., Cox, P., Owings, A.D. and Valverde, R.A. (1998) Research on disease of ornamental plants. Luisiana Agriculture, 41, 4-5.
- Bellardi, M.G., Rubies, A. and Cerato, C. (2001) Identification of an isolate of potato virus—Y tuber necrotic strain of clary sage (Salvia sclarea); Plant Dis. (St. Paul. Minn). American Phytopathological Society, 85, 1207
- van Regenmortel, M.H.V., Fauquet, C.M., Bishop, D.H.L., Carstens, E.B., Estes, M.K., Lemon, S.M., Maniloff, J., Mayo, M.A., McGeoch, D.J., Pringle, C.R. and Wickner R.B. (2000) Virus taxonomy classification and nomenclature of viruses. The 7th Report of International Committee on Taxonomy of Viruses, Academic Press, California.
- Palukaitis, P., Roossinck, M., Dietzgen, R.G. and Francki, R.I.B. (1992) Cucumber Mosaic Virus. Advances in Virus Research, 41, 281-348. doi:10.1016/S0065-3527(08)60039-1
- Suzuki, M., Kuwata, S., Kataoka, J., Masuta, C., Nitta, N. and Takanami, Y. (1991) Functional analysis of deletion mutants of Cucumber Mosaic Virus RNA 3 using an in vitro transcription system. Virology, 183, 106-113. doi:10.1016/0042-6822(91)90123-S
- Wahyuni, W.S., Dietzen, R.G., Handa, K. and Francki, R.I.B. (1992) Serological and biological variation between and within subgroup I and II strains of Cucumber mosaic virus. Plant Pathology, 41, 282-297. doi:10.1111/j.1365-3059.1992.tb02350.x
- Hu, J.S., Li, H.P., Berry, K., Wang, M. and Jordan, R. (1995) Comparison of dot blot ELISA and RT-PCR assays for detection of two cucumber mosaic virus isolates infecting banana in Hawaii. Plant Disease, 79, 202-206. doi:10.1094/PD-79-0902
- Ilardi, V., Mazzei, M., Loreti, S., Tomassoli, L. and Barba, M. (1995) Biomolecular and serological methods to identify strains of cucumber mosaic cucumovirus on tomato. EPPO Bulletin, 25, 321-327. doi:10.1111/j.1365-2338.1995.tb01473.x
- Owens, J. and Palukaitis, P. (1988) Characterization of Cucumber mosaic virus. I. Molecular heterogeneity mapping of RNA 3 in eight CMV strains. Virology, 69, 496- 502.
- Owens, J., Shintaku, M., Aeschleman, P., Tahar, S.F. and Palukaitis, P. (1990) Nucleotide sequence and evolutionary relationships of Cucumber Mosaic Virus (CMV) strains. CMV RNA 3. Journal of General Virology, 71, 2243-2249. doi:10.1099/0022-1317-71-10-2243
- Szilassy, D., Sala’nki, K. and Bala’zs, E. (1999) Molecular evidence for the existence of two distinct subgroups in Cucumber Mosaic Virus. Virus Genes, 18, 221-227. doi:10.1023/A:1008016202128
- Rizos, H., Gunn, L.V., Pares, R.D. and Gillings, M.R. (1992) Differentiation of cucumber mosaic virus isolates using polymerase chain reaction. Journal of General Virology, 73, 2099-2103. doi:10.1099/0022-1317-73-8-2099
- Sialer, M.M., Cillo, F., Barbarossa, L. and Gallitelli, D. (1999) Differentiation of Cucumber Mosaic Virus subgroups by RT-PCR, RFLP. Journal of Plant Pathology, 81, 145-148.
- Palukaitis, P. and Zaitlin, M. (1997) Replicase-mediated resistance to plant virus diseases. Advances in Virus Research, 48, 349-377. doi:10.1016/S0065-3527(08)60292-4
- Roossinck, M.J. (2002) Evolutionary history of Cucumber mosaic virus deduced by phylogenetic analysis. Journal of Virology, 76, 3382-3387. doi:10.1128/JVI.76.7.3382-3387.2002
- Eiras, M., Boari, A.J., Colariccio, A., Chaves, A.L.R., Briones, M.R.S., Figueira, A.R. and Harakava, R. (2004) Characterization of isolates of the cucumovirus Cucumber Mosaic Virus present in Brazil. Journal of Plant Pathology, 86, 61-69.
- Singh, D., Naqvi, Q.A. and Garg, I.D. (1999) A strain of Cucumber Mosaic Virus causing mosaic in marigold in India. Indian Phytopathology, 52, 114-117.
- Walkey, D.G.A. (1985) Applied plant virorogy. William Heinmann Ltd., London.
- Christie, S.R., Purciful, D.E., Grawford, W.E. and Ahmad, N.A. (1987) Electron microscopy of negatively stained clarified virus concentrates obtained from small tissue samples with appendices of negative staining techniques. Institute of Food and Agriculture Science, University of Florida Technical Bulletin, Gainesville.
- Garg, I.D. and Khurana, S.M.P. (1992) Factors influencing immune electron microscopy of flexuous potato viruses. Acta Virologica, 36, 435-442.
- Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular coning: A laboratory manual. Cold Spring Harbor Laboratory, New York.
- Srivastava, A., Chandra, G. and Raj, S.K. (2004) Coat protein and movement protein gene based molecular characterization of Amaranthus strain of Cucumber Mosaic Virus. Acta Virologica, 48, 229-239.
- Thompson, J.D., Higgins, D.G., Gibson, T.J. and Clustal, W. (1994) Improving the sensitivity of progressive multiple sequence alignment through sequence weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22, 4673-4680. doi:10.1093/nar/22.22.4673
- Tamura, K., Dudley, J., Nei, M. and Kumar, S. (2007) Mega4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596-1599. doi:10.1093/molbev/msm092
- Wang, X.F., Pei, M.Y., Ma, D.F. and Tian, W.H. (1985) Studies on scarlet sage mosaic disease. Acta Phytopathologica Sinica, 15, 36-46.
- Hu, J.S., Li, H.P., Barry, K., Wang, M. and Jordan, R. (1995) Comparision of dot.blot ELISA and RT-PCR assay for detection of two cucumber mosaic isolates infecting banana in Hawaii. Plant Disease, 79, 902-906. doi:10.1094/PD-79-0902
- Carrere, I., Tepfer, M. and Jacquemond, M. (1999) Recombinants of Cucumber Mosaic Virus (CMV): determinants of host range and symptomatology. Archives of Virology, 114, 365-379.
- Takeshita, M., Suzuki, M. and Takanami, Y. (2001) Combinations of amino acids in the 3a protein and the coat protein of cucumber mosaic virus determines symptom expression and virus spread in bottle gourd. Archives of Virology, 146, 697-711. doi:10.1007/s007050170140

