Food and Nutrition Sciences
Vol.5 No.13(2014), Article
ID:47652,9
pages
DOI:10.4236/fns.2014.513125
Functional Characterization of Bifidobacteria of Human Origin: A Case Study by the Students of Food Science and Technology of the University of Foggia (Southern Italy)
Emanuela Ciuffreda, Angino Veronica, Antonietta Cifelli, Rossana Foti, Rosaria Ilenia Forte, Fabio Graziani, Andrea Giuseppe Longo, Angela Longo, Valeria Maglia, Emilio Francesco Ricciardi, Alessandra Sabatino, Annamaria Tomaiuolo, Maria Rosaria Corbo, Milena Sinigaglia, Antonio Bevilacqua*
Department of the Science of Agriculture, Food and Environment (SAFE), University of Foggia, Foggia, Italy
Email: *antonio.bevilacqua@unifg.it, *abevi@libero.it
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 10 April 2014; revised 20 May 2014; accepted 2 June 2014
ABSTRACT
The aim of this paper was to study the potential technological and probiotic properties of bifidobacteria isolated from human feces. Bifidobacteria, naturally present in the dominant colonic microbiota, represent up to 25% of the cultivable faecal bacteria in adults and 80% in infants. Bifidobacteria have been shown to adhere and colonize in high numbers different types of cultured intestinal epithelial cells; moreover some authors reported that some strains are able to stabilize the intestinal microbiota during and after antibiotic therapy, modulate the immune system, protecting against chemically induced intestinal inflammation and reducing symptoms of colitis. Eight isolates of bifidobacteria were studied to assess their technological and probiotic traits; the technological characterization relied on the assessment of enzymatic activities (proteolytic and lipolytic activity), growth under various conditions (pH, temperature and addition of salt), acidifying ability and metabolism (arginine deamination, esculin, esculin hydrolysis and citrate metabolism). The study of the probiotic characteristics focused on the evaluation of the survival at low pH and with bile salts added, antibiotic resistance, and hydrophobic properties. As a result of this process, two promising strains were selected for further studies.
Keywords:Bifidobacteria, Technology, Probiotic Traits, Functional Characterization

1. Introduction
Microorganisms play a very important role in human health and nutrition; they are involved in the production of various biomolecules, fermentation of food and also reside symbiotically in gut benefiting the host. Various microorganisms have traditionally been employed for manufacture of fermented milks and cheeses and bifidobacteria are well known for their role as probiotics for dairy products [1] .
Bifidobacteria are Gram positive, non spore forming, non motile rods with high GC content; they are anaerobic to aerotolerant and are generally catalase negative. They are able to ferment glucose to lactic and acetic acids through the F6PPK-pathway (fructose-6-phosphate phosphoketolase) [2] . Bifidobacteria have a long standing history of safe use in fermented food products and possess GRAS (Generally recognized as safe) and QPS (Qualified presumption of safety) status [3] .
Some bifidobacteria strains have been shown to adhere and also to colonize in high numbers different types of cultured intestinal epithelial cells [4] . Some authors reported that some strains are able to alleviate chronic intestinal disorders, such as inflammatory bowel disease, traveler’s diarrhea, colitis, Crohn’s disease, and antibiotic-associated diarrhea [5] -[7] is increasingly being documented.
A new kind of starter cultures is the functional starters, i.e. microorganisms acting at the same time as probiotic and starter [8] . Functional starter cultures are used for the improvement of aroma, to obtain safe products because of their ability to produce bacteriocins, for their ability to enrich food matrix with micronutrients. Some reports are available on the use of lactobacilli as functional starter cultures; but to the best of our knowledge few data are available on the bifidobacteria, therefore the main topic of this research was to study the technological and probiotic characteristics of some strains of bifidobacteria. In particular, we focused on eight strains isolated from human feces, assessing:
1) the growth at different temperatures, NaCl amounts and pH values;
2) the acidifying ability;
3) the metabolism (arginine deamination, esculin hydrolysis, proteolytic and lipolytic activity, citrate metabolism);
4) the survival at low pH and in presence of bile salts;
5) the hydrophobic properties;
6) the antibiotic resistance.
2. Materials and Methods
2.1. Microorganisms
This research focused on 8 isolates of human origin; the microorganisms, labeled with a numeric code were stored at −20˚C in MRS broth (Oxoid, Milan, Italy), added with 33% of sterile glycerol (J.T. Baker, Milan). Before each assay the strain were grown in MRS, incubated at 37˚C for 24 h.
2.2. Arginine Deamination
Arginine deamination was assessed in the medium by Abd-El-Malek buffered to pH 7.
The composition of the substrate is shown below:
• tryptone 5 g/l;
• yeast extract 2.5 g/l;
• glucose 0.5 g/l;
• K2HPO4 2 g/l;
• arginine hydrochloride 3 g/l.
The medium (aliquots of 5 ml) was inoculated to 7 log cfu/ml with each strain separately; then the samples were incubated for 4 days at 37˚C. After adding the Nessler reagent, if the color of the samples turns to orange, the test was considered positive.
2.3. Aesculin Hydrolysis
The test was performed in MRS broth, containing 2 g/l of aesculin (Sigma-Aldrich, Milan), 1 g/l of ferric ammonium citrate (1 g/l) (C. Erba, Milan) and buffered to pH 6.5. After the inoculation of bifidobacteria (ca. 7 log cfu/ml), the samples were incubated at 37˚C for 3 - 4 days; a black color of the medium indicated aesculin hydrolysis.
2.4. Use of Citrate
The strains were streaked onto the surface of Simon's Citrate Agar (Oxoid), incubated for 3 - 5 days at 37˚C. The test was considered positive if the color of the medium turned from green to blue.
2.5. Proteolytic Activity
The bacteria were inoculated by spot (20 μl of a cell culture at 9 log cfu/ml) onto the surface of Plate Count Agar + 5% of skim milk; the plates were incubated at 37˚C for 3 - 5 days. A clear halo surrounding the colony was the sign of a proteolytic activity.
2.6. Lipolytic Activity
The strains were inoculated by spot (20 μl of a cell culture at 9 log cfu/ml) onto the surface of Plate Count Agar + 2% of tributyrin (Sigma-Aldrich); the plates were incubated at 37˚C for 3 - 5 days. A clear halo surrounding the colony was the sign of a proteolytic activity.
2.7. Technological Characterization
The strains were inoculated in MRS broth (ca. 5 log cfu/ml), containing different amounts of NaCl (2%, 4%, or 6%), adjusted to acidic or alkaline pH (4, 5, and 8) or incubate at various temperatures (15˚C, 25˚C, and 44˚C). Aliquots of not-modified MRS, inoculated with bifidobacteria and incubated at 37˚C, were used as controls.
Microbial growth was evaluated after 24 and 48 h as absorbance measurement at 600 nm; the analyses were performed in duplicate and the results modeled as Growth Index (GI) [9] :

where Abss is the absorbance of the samples and Absc the optical density of the control, at time t.
2.8. Acidification
Aliquots of MRS, MRS + 4% NaCl, and skim milk were inoculated to 7 log cfu/ml with each strain separately; the strain were stored at 15˚C, 25˚C, 37˚C and 44˚C. The pH was evaluated immediately after the inoculation and after 24 h through a pH meter Crison (Crison Instruments, Barcelona, Spain).
All the experiments were performed in duplicate and the data modeled as decrease of the pH referred to the beginning of the assay.
2.9. Hydrophobicity
The bacterial cell surface hydrophobicity was assessed by measuring the microbial adhesion to hydrocarbons using the procedure described by Crow et al. [10] and Bautista-Gallego et al. [11] . Briefly, cells at the stationary phase were centrifuged (10,000 ×g, 5 min).
The resulting pellet was washed twice in phosphate buffered saline solution (PBS), re-suspended in 3 ml of 0.1 mol·l−1 KNO3 (final concentration of cells was ca. 108 cfu/ml) and the absorbance at 600 nm was measured (A0). One milliliter of xylene (Merck, Darmstadt, Germany) was then added to the cell suspension to form a two-phase system.
After a 10 min pre-incubation at room temperature, the two-phase system was mixed by vortexing for 2 min. Then, the water and xylene phases were separated by incubation for 20 min at room temperature.
The aqueous phase was carefully removed and the absorbance at 600 nm was measured (A1). The percentage of the cell surface hydrophobicity (HI) was calculated using the formula HI = (1 − A1/A0) × 100.
2.10. Survival at pH 2.5 and with Bile Salts Added
The survival into the gut was assessed in a model system, consisting of acidified distilled water or distilled water with 0.3% bile salts (Oxoid). The strains were grown and washed as reported above, then inoculated in the following samples (ca. 107 cfu/ml): a) distilled water acidified to pH 2.5; b) distilled water containing bile salts (0.3%); c) distilled water (control).
The samples were incubated at 37˚C for 3 h; then, they were serially diluted in a saline solution (0.9% NaCl) and plated onto MRS agar (37˚C for 48 - 72 h).
2.11. Antibiotic Resistance
This assay was performed through a modified version of the E-test, using the antibiotic strips produced by Liofilchem (Roseto degli Abruzzi, Italy). Bifidobacteria were streaked onto the surface of MRS agar through a sterile swab; therafter, the strip containing the antibiotic was placed onto the surface of the plates.
The plates were incubated at 37˚C for 24 h; a clear halo was the sign of the susceptibility of the microbial target to the tested antibiotic. The Minimal Inhibitory Concentration of each antibiotic was evaluated as suggested by the producer. The following antibiotics were used in this assay (the values in the brackets are the range of antibiotic concentrations on the strip):
1) Ampicillin (0.016 - 256 µg).
2) Ciprofloxacin (0.002 - 32 µg).
3) Chloramphenicol (0.016 - 256 µg).
4) Clarithromycin (0.016 - 256 µg).
5) Erythromycin (0.016 - 256 µg).
6) Gentamycin (0.064 - 1024 µg).
7) Streptomycin (0.064 - 1024 µg).
8) Tetracycline (0.016 - 256 µg).
9) Trimethoprim (0.002 - 32 µg).
10) Vancomycin (0.016 - 256 µg).
2.12. Biochemical Characterization and Phenotypic Identification
The strains were identified through the miniaturized system API 20 A (Marcy L’Etoile, France). the fermentative profile was regarded as acceptable if the accordance between the profile of each strain and those present in the software was at least 95%.
2.13. Statistical Analysis
The experiments were performed in duplicate over two different batches; thereafter the data of the quantitative assays (survival at pH 2.5 and with bile salts, acidification in skim milk at 37˚C and 45˚C, growth with salt, at different pHs and temperatures, hydrophobicity) were used as input variables to run a Principal Component Analysis through the software Statistica for Windows, version 10.0 (Statsoft, Tulsa, Okhla.).
3. Results and Discussion
The selection of lactic acid bacteria as suitable starters cultures/probiotic is a complex process, involving the evaluation of some technological performances and desired metabolic traits, along with the study of the probiotic properties; this paper proposes a simple approach, based upon some primary technological tools (growth under low and relatively high temperatures, resistance to salt, acidification and ability to overcome stressful conditions) and probiotic traits (survival at pH 2.5 and with bile salts added, ability to adhere to the intestinal mucosa, antibiotic resistance).
3.1. Phenotypic Trait and Technological Characterization
The strains were identified through the phenotypic approach as Bifidobacterium spp. (data not shown). Table 1
Table 1. Phenotypic traits of bifidobacteria.
shows the phenotypic traits of bifidobacteria; none of the tested strains was able to hydrolyze aesculin. On the other hand all the isolates showed a kind of proteolytic activity (from medium to strong). The other traits were strain-dependent.
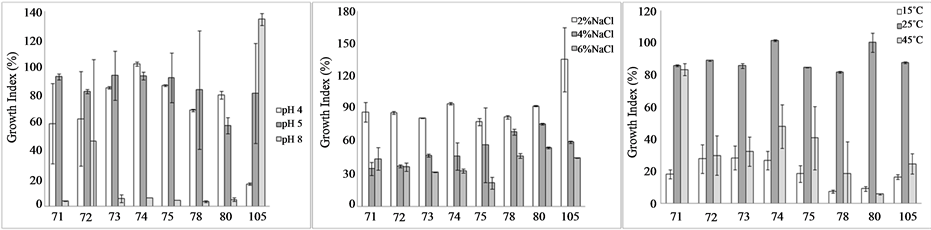
The results of the growth assays are shown in the Figures 1(a)-(c); the figures show the Growth index as a function of pH, salt and temperature. This approach was proposed by Bevilacqua et al. [9] for the characterization of yeasts isolated from table olives, later used for some kinds of bacteria (enterobacteria or lactic acid bacteria) [12] ; the numerical range of GI can be used to state of the effect of the factor (inhibition, partial inhibition or no effect). Namely, when GI is <25%, the microorganisms are completely inhibited; on the other hand, a GI in the range 25% - 75% stands for a partial inhibition.
Concerning the effect of pH (Figure 1(a)), the strain 104 was completely inhibited at pH 4, whilst the isolates 71 and 72 (GI ca. 60%) were partially affected. Under alkaline conditions, all the strains were inhibited except for the isolates 72 and 104; in particular, the strain 72 was partially affected, as it experienced a GI of ca. 40%, while the GI of the isolate 104 was 137%, thus suggesting a kind of benefit at this pH.
As expected, salt affected significantly microbial growth and its effect relied upon the amount, with a GI ranging from 15% to 48% in presence of 6% of NaCl (Figure 1(b)).
The effect of the temperature was strictly strain-dependent; generally, at 25˚C the G was >80%, thus suggesting that at this temperature the growth was quite similar to that recovered at 37˚C. Moreover, the strains appeared inhibited (partially or completely) both at 15˚C and 45˚C (Figure 1(c)); only the strain 72 showed a GI of 80% at 45˚C.
The last experiment of the technological characterization focused on the acidification exerted by bifidobacteria in a model system; the decrease of the pH (Figure 2) was a function of the temperature and the kind of the medium. In not-modified MRS bifidobacteria decreased slightly the pH at 15˚C (by 0.4 - 0.6); an increase of the temperature caused a significant increase of ΔpH (1.7 - 2.2). In MRS added with salt, bifidobacteria were completely inhibited at 15˚C and slightly affected at 25˚C.
Finally, in skim milk the temperature exerted a stronger effect on some strains (73, 74, 78, 80, and 105), as they experienced a significant ΔpH at 37˚C, but not at 15˚C and 25˚C.
3.2. Probiotic Properties
Figure 3 shows the HI (hydrophobic index) of bifidobacteria; the highest index was found for the isolate 80 (7.40%), whilst a certain degree of hydrophobicity was recovered also for the isolates 73 and 74 (2.47 and 4.37%, respectively). The other bifidobacteria did not possess this trait.
Concerning the resistance to the conditions of the gut, all the strains were not affected by bile salts, whilst the effect of the pH was very strong for the isolates 71 and 72, which were reduced by 7.3 - 8 log cfu/ml (Figure 4).
Table 2 shows MIC values of the antibiotics; the targets were resistant to ciprofloxacin, trimethoprim and vancomycin, whilst gentamycin and streptomycin showed the highest MICs (respectively 57 and 333.71 µg).
Concerning the probiotic aptitudes, the survival into the gut and the intestinal adhesion are two important prerequisites to maintain the functionality within the host [13] ; the binding to the mucosa is a multistep process,
 (a) (b) (c)
(a) (b) (c)
Figure 1. Growth Index of bifidobacteria in MRS broth after 24 h, as a function of pH, NaCl and storage temperature. Mean values ± standard deviation.
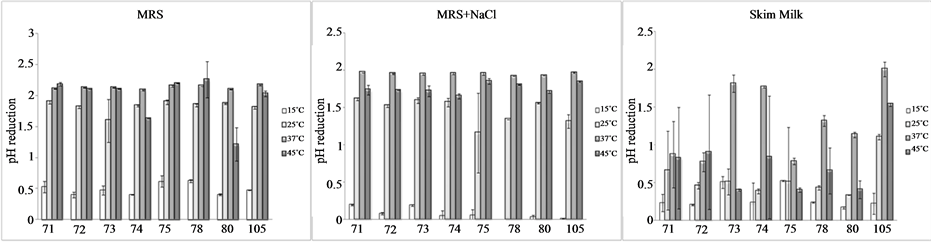
Figure 2. Reduction of pH by bifidobacteria after 24 h in MRS, MRS + 4% NaCl and skim milk. Mean values ± standard deviation.

Figure 3. Hydrophobic index of bifidobacteria towards xylene. Mean values.
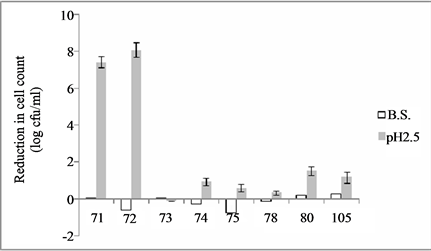
Figure 4. Survival of bifidobacteria at pH 2.5 and with bile salts added (0.3%) (B.S.). The data are reported as decrease of cell count referred to the inoculum; mean values ± standard deviation.
Table 2. Minimal Inhibitory Concentration (µg) of the antibiotics towards bifidobacteria.
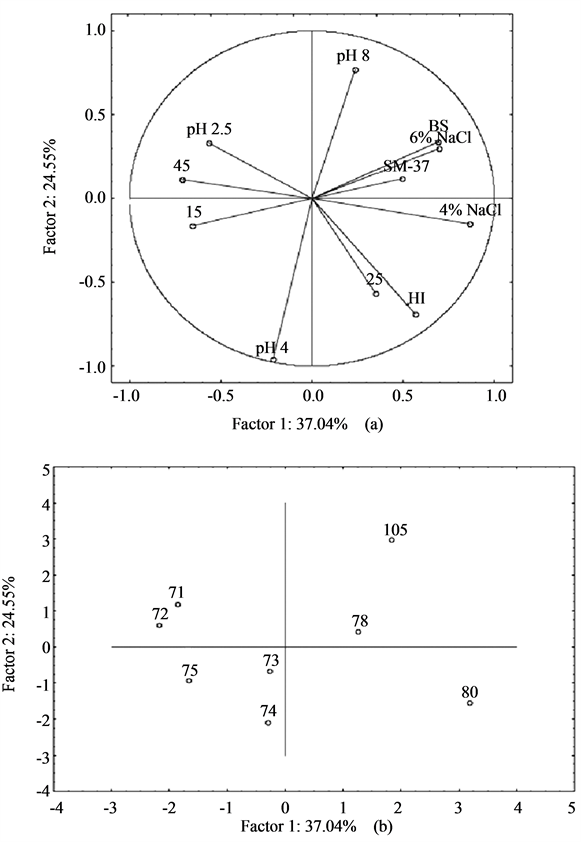
Figure 5. Principal Component Analysis on the technological and probiotic traits of bifidobacteria. (a) Projection of the variables; (b) Strain distribution in the factorial space. B.S. and pH 2.5, decrease of cell count after 3 h in distilled water acidified to pH 2.5 or containing 0.3% of bile salts; HI, hydrophobic index; SM-37, acidification in skim milk at 37˚C; pH 4, 5, 8; NaCl 4, 6%; 15, 25, 45: Growth Index after 24 h at pH 4, 5 or 8, or with NaCl (4% and 6%) or at 15˚C, 25˚C and 45˚C.
which involves both electrostatic interactions and hydrophobic forces [14] . The classical assay for this trait is the adhesion to intestinal cell lines under laboratory conditions; however, some authors [11] reported that the strains able to adhere to cell monolayers (Caco, IPEC-J2) possess hydrophobicity characteristics as determined by the assays with hydrocarbons. Therefore, the hydrophobicity was used as an indirect and screening tool to test the potentiality of the isolates to bind to the intestinal mucosa.
Concerning the antibiotic resistance, the recent guidelines of the European Food Safety Authority state that probiotic should not transfer the genes of the resistance to other microorganisms, thus it would preferable to have probiotics sensitive to the antibiotics or possessing a cromosomial resistance [15] .
3.3. Strain Selection
In the last step of the research a multivariate approach was used to select the most promising strain for a possible in vivo validation; some authors proposed in the past different kinds of multivariate approaches to select suitable starter cultures (Principal Component Analysis-PCA, Cluster Analysis etc.) [16] [17] .
The Cluster analysis is a tool for the molecular biology to point out and highlight biotypes and/or subspecies, whilst the Principal Component Analysis has two main benefits: strain grouping and pointing out the variables responsible for strain differences [8] . In this paper we used PCA.
Figure 5(a) shows the projection of the variables in the factorial space; the technological and the probiotic traits were not related, thus it was not possible to select the best strain; on the other hand, we focused on some promising strains, like the isolates 80 and 105, both resistant to the pH of the stomach (2.5) and to NaCl and able to decrease the pH of skim milk. The strain 80 was also characterized by a high Hydrophobic Index.
A possible limit of these two strains could be the relatively low Growth Index at 15˚C and 45˚C, thus suggesting that they could exert their metabolic potential under strictly controlled thermal profile.
4. Note by Supervisor
This paper was written by the students of Food Science and Technology degree, University of Foggia, as a kind of final examination. Students attended the lessons in the lab and each lesson consisted of 3 - 4 different parts: theoretical introduction and critical reading of the protocols; practical experiment in the laboratory; data collection and analysis; critical discussion of the data.
After a final focus group, each student was requested to write a part (mainly the Materials and Methods and the Results of a single experiment); thereafter, the contributions were collected, corrected by the supervisor and organized as a paper.
References
- Ishizeki, S., Sugita, M., Takata, M. and Yaeshima, T. (2013) Effect of Administration of Bifidobacteria on Intestinal Microbiota in Low-Birth-Weight Infants and Transition of Administered Bifidobacteria: A Comparison between One-Species and Three-Species Administration. Anaerobe, 23, 38-44. http://dx.doi.org/10.1016/j.anaerobe.2013.08.002
- Martinez, F.A.C., Balciunas, E.M., Converti, A., Cotter, P.D. and de Souza Oliveira, R.P. (2013) Bacteriocin Production by Bifidobacterium spp. A Review. Biotechnology Advances, 31, 482-488. http://dx.doi.org/10.1016/j.biotechadv.2013.01.010
- European Food Safety Authority (EFSA) (2007) Introduction of a Qualified Presumption of Safety (QPS) Approach for Assessment of Selected Microorganisms Referred to EFS (Question No EFSA-Q-2005-293). The EFSA Journal, 587, 1-16.
- Wang, M., Ahrnes, S., Jeppsson, B. and Molin, G. (2005) Comparison of Bacterial Diversity along the Human Intestinal Tract by Direct Cloning and Sequencing of 16S rRNA Genes. FEMS Microbiology Ecology, 54, 219-231. http://dx.doi.org/10.1016/j.femsec.2005.03.012
- Kaur, I.P., Kuhad, A., Garg, A. and Chopra, K. (2009) Probiotics: Delineation of Prophylactic and Therapeutic Benefits . Journal of Medicinal Food, 12, 219-235. http://dx.doi.org/10.1089/jmf.2007.0544
- Marteau, P.R. (2002) Probiotics in Clinical Conditions. Clinical Reviews in Allergy Immunology, 22, 255-273. http://dx.doi.org/10.1007/s12016-002-0011-0
- Ohland, C.L. and Macnaughton, W.K. (2010) Probiotic Bacteria and Intestinal Epithelial Barrier Function. American Journal of Physiology-Gastrointestinal and Liver Physiology, 298, G807-G819. http://dx.doi.org/10.1152/ajpgi.00243.2009
- Bevilacqua, A., Cagnazzo, M.T., Caldarola, C., Ciuffreda, E., Dragano, A.R., Franchino, S., Lauriola, R., Pacifico, A., Corbo, M.R. and Sinigaglia, M. (2012) Bifidobacteria as Potential Functional Starter Cultures: A Case Study by MSc Students in Food Science and Technology (University of Foggia, Southern Italy). Food and Nutrition Sciences, 3, 55-63.
- Bevilacqua, A., Perricone, M., Cannarsi, M., Corbo, M.R. and Sinigaglia, M. (2009) Technological and Spoiling Characteristics of the Yeast Microflora Isolated from Bella di Cerignola Table Olives. International Journal of Food Science and Technology, 44, 2198-2207. http://dx.doi.org/10.1111/j.1365-2621.2009.02060.x
- Crow, V., Gopal, P. and Wicken, A. (1995) Cell Surface Differences of Lactococcal Strains. International Dairy Journal, 5, 45-68. http://dx.doi.org/10.1016/0958-6946(94)P1598-8
- Bautista-Gallego, J., Arroyo-López, F.N., Rantsiou, K., Jiménez-Díaz, R., Garrido-Fernández, A. and Cocolin, L. (2013) Screening of Lactic Acid Bacteria Isolated from Fermented Table Olives with Probiotic Potential. Food Research International, 50, 135-142. http://dx.doi.org/10.1016/j.foodres.2012.10.004
- Bevilacqua, A., Altieri, C., Corbo, M.R., Sinigaglia, M. and Ouoba, L.I.I. (2010) Characterization of Lactic Acid Bacteria Isolated from Italian Bella di Cerignola Table Olives: Selection of Potential Multifunctional Starter Cultures. Journal of Food Science, 75, M536-M544.
- van der Aa Kuhle, A., Skovgaardb, K. and Jespersen, L. (2005) In Vitro Screening of Probiotic Properties of Saccharomyces cerevisiae var. boulardii and Food-Borne Saccharomyces Cerevisiae Strains. International Journal of Food Microbiology, 101, 29-39. http://dx.doi.org/10.1016/j.ijfoodmicro.2004.10.039
- Warne Zoueki, C., Ghoshal, S. and Tufenkji, N. (2010) Bacterial Adhesion to Hydrocarbons: Role of Asphaltenes and Resins. Colloids and Surfaces B: Biointerfaces, 79, 219-226. http://dx.doi.org/10.1016/j.colsurfb.2010.03.054
- Sanders, M.E., Akkermans, L.M.A., Haller, D., Hammerman, C., Heimbach, J., Hörmannsperger, G., Huys, G., Levy, D.D., Lutgendorff, F., Mack, D., Phothirath, P., Solano-Aguilar, G. and Vaughan, E. (2010) Safety Assessment of Probiotics for Human Use. Gut Microbes, 1, 164-185. http://dx.doi.org/10.4161/gmic.1.3.12127
- Rodr1guez-Gomez, F., Romero-Gil, V., Bautista-Gallego, J., Garrido-Fernandez, A. and Arroyo-Lopez, F.N. (2012) Multivariate Analysis to Discriminate Yeast Strains with Technological Applications in Table Olive Processing. World Journal of Microbiology and Biotechnology, 28, 1761-1770.
- Bevilacqua, A., Beneduce, L., Sinigaglia, M. and Corbo, M.R. (2013) Selection of Yeasts as Starter Cultures for Table Olives. Journal of Food Science, 78, M742-M751.
NOTES

*Corresponding author.


