Crystal Structure Theory and Applications
Vol.04 No.04(2015), Article ID:61428,6 pages
10.4236/csta.2015.44006
Single-Chain Expression and Crystallization of an Antigenic C-Terminus in Complex with the Regulatory Domain of ER Aminopeptidase 1
Lufei Sui1, Amit Gandhi1,2, Hwai-Chen Guo1*
1Department of Biological Sciences, University of Massachusetts Lowell, Lowell, MA, USA
2Harvard Medical School, Boston, MA, USA

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 2 October 2015; accepted 21 November 2015; published 24 published 2015

ABSTRACT
Human endoplasmic reticulum aminopeptidase 1 (ERAP1) is one of two ER luminal aminopeptidases that participate in the final processing of peptide precursors and generates the N-termini of the MHC class I-restricted epitopes. In order to investigate the interactions of its binding site with substrate peptides, X-ray crystallographic analyses have been carried out to study structures of ERAP1 regulatory (ERAP1_R) domain in complex with antigenic peptides. Single-chain bimodular constructs with various antigenic peptides linked to the C-terminal end of ERAP1_R domain are designed to facilitate crystallization process of these complexes. These recombinant proteins have been purified and crystalized, and x-ray diffraction data of one crystal have been processed to a resolution of 2.8 Å. The crystal belongs to the space group P21, with unit cell parameters a =64.2, b = 66.8, c = 66.3 Å, β = 110.2˚. A Refmac-refined omit map reveals a clear density for the antigenic peptide’s carboxylate-end that is in contact with the ERAP1 regulatory domain of neighboring molecule. Thus the single-chain bimodular constructs have provided an expedited approach to study sequence-specific interactions between the ERAP1 regulatory domain and antigen peptide’s C-terminal ends.
Keywords:
Endoplasmic Reticulum Aminopeptidase 1 (ERAP1), ERAP1 Regulatory Domain, Antigen Presentation, X-Ray Crystallography

1. Introduction
During antigen processing in the human immune system, precursor peptides are transported into endoplasmic reticulum (ER) via transporter associated with antigen processing (TAP). ERAP1 then cleaves N terminal extended residues of the precursors inside the ER to generate mature antigens [1] [2] . These mature antigens are subsequently passed on to major histocompatibility complex class I (MHC1) proteins for presentation on the cell surface in order for cytotoxic (CD8+) T cells to recognize [3] . However, due to the size and shape of the binding pockets, MHC1 proteins have a length-restriction on mature antigen to be 8 to 10 residues [4] [5] . Therefore, precursor peptides need to be precisely trimmed by ERAP1 before they can be loaded onto MHC1 molecules for cell surveillance.
ERAP1 has both length and residue preference for the substrates [6] . Its catalytic activity is triggered by a unique allosteric mechanism, dubbed molecular ruler mechanism that controls the substrate length and inspects the nature of the C-terminal amino acid 9 - 16 residues away from the N-terminal catalytic site [3] . The length preference of ERAP1 corresponds to the lengths of N-extended antigenic precursors transported by TAP. According to this molecular ruler mechanism, it can accommodate peptides with 9 to 16 amino acid long and preferably with a hydrophobic C-terminal residue. In addition, other studies reveal that the internal sequence of substrate peptides also affects the trimming efficiency of ERAP1 [7] . Crystal structures of ERAP1 show that there is likely a conformational change from open form to closed form, which is associated with substrate binding and activation of catalytic site [6] [8] . Open form of ERAP1 is presumed to be inactive without a substrate. Binding of substrate to the regulatory site (binding groove) is proposed to convert ERAP1 to a closed form, in which the N-terminal residue of the substrate is cleaved at the catalytic site [9] . Due to the length preference of ERAP1, it precisely cleaves the N-extended peptides to eight or nine residues, while further trimmings slow down or cease completely [3] . In this way, it produces epitopes with optimal length that are presented by most MHC1 molecules. Thus, the substrate specificity of ERAP1 is of great immunological importance, considering its influence on the selectivity of peptides for trimming and therefore their availability for MHC1 presentation.
We previously reported a structure of ERAP1 regulatory domain (aa 529 - 941) in complex with a carboxyl-terminal hexa-histidine tag [9] . It reveals the binding mechanism of ERAP1 regulatory domain and explains how ERAP1 can monitor the substrate length at substrate’s C-terminus that is located about 30 Å away from the N-terminal catalytic site.
Taking advantage of the complex formation between the ERAP1 regulatory domain (ERAP1_R) and the C-terminal His tag during crystallization, we have designed single-chain constructs to facilitate purification and crystallization of ERAP1_R/peptide complexes. To this end, the His6 tag at the C-terminus of the ERAP1_R in the previous construct is replaced with peptide sequences derived from the C-terminal ends of a few natural antigens. The new recombinant proteins have been purified, and crystals are obtained. Preliminary crystallographic analyses suggest that new antigen-tagged proteins form the same crystal form, suggesting the same inter-molecular contacts in the crystals that will allow high-resolution crystallographic studies of sequence-specific interactions between the ERAP1 regulatory domain and antigen peptide’s C-terminal ends.
2. Materials and Methods
2.1. Protein Expression and Purification
Baculovirus vectors carrying inserts encoding for ERAP1 regulatory domain (ERAP1_R) with various peptide sequences at C-terminal ends (Table 1) were constructed with a N-terminal hexa-histidine tag according to the protocols of the manufacturer (Invitrogen). The presence of ERAP1 protein and the integrity of the purified recombinant bacmid DNA were verified by PCR and sequencing. To express the protein, the bacmid DNA was transfected into Sf9 insect cells according to the manufacturer’s protocols. The protein was expressed by adding the P3 recombinant viral stock into Sf9 insect cells with an MOI of 0.8 pfu/cell, and harvested 54 hours after infection. Protein expression was confirmed by western blot using primary antibody against the hexa-histidine tag. Cell pellet was re-suspended in 50 mM NaH2PO4, pH 8.0, 300 mM NaCl and 10 mM imidazole, and lysed by freeze-thaw cycles and sonication. The supernatant was loaded onto a Ni-NTA column and washed several times with 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl, and 10 - 30 mM imidazole. The protein was then eluted with 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl, and 400 mM imidazole. Glycerol was added to the eluted solution to a final concentration of 16% (v/v), and then the concentrated protein was further purified through a Superdex 200 gel filtration column (Amersham Pharmacia) by FPLC system with a buffer containing 10 mM Tris, pH 7.5, 10 mM NaCl. A single peak for ERAP1_R domain was collected and the protein was concentrated to 5 mg/ml for crystallization.
2.2. Crystallization
Hanging-drop vapor diffusion technique was used for initial crystallization screening at 4˚C (Table 2). Micro-seeding method was used to improve crystal quality. The best looking crystal was formed above a well solution containing 100 mM Tris, pH 8.5 and 16% PEG8000 at 4˚C in 7 days.
2.3. Diffraction Data Collection and Processing
For data collection, the crystal was cryoprotected in solution containing 100 mM Tris-HCl buffer (pH 8.0) and 30% glycerol. X-ray data were collected using the beamline X29 at National Synchrotron Light Source (NSLS) - Brookhaven. The data were processed with the Mosflm29 and the CCP4 suite30.
3. Results and Discussion
Various peptides with 6 - 10 different amino acids were designed (Table 1) based on a few natural antigenic peptide sequences: an ovalbumin epitope SIINFEKL (underlined sequence is included in one construct, [3] ), a HSP-86 epitope RRIKEIVKKH [10] , a HSP-90 epitope AEDKENYKKF [11] , and an optimized substrate sequence based on library screening LVAFKARKF [7] . In addition, one construct contains a poly-Aalanine linker, AAAAFKARKF-COOH, was designed to test length flexibility during crystallization to form intermolecular peptide/ERAP1_R complex. These peptide sequences were added to the C terminal end of ERAP1 regulatory domain ERAP1_R.
All the recombinant proteins with different antigenic peptides were successfully expressed in insect cells and the expression yield was optimized and analyzed by western blot. After Ni-NTA affinity chromatography, protein elution fractions were collected and concentrated for size exclusion chromatography. Based on Superdex 200 gel filtration standard, peak fractions around the elution volume of 15 ml was collected and further concentrated up to 5 mg/ml with at least 95% purity as analyzed by SDS PAGE (Figure 1).
After initial screening by hanging drop vapor diffusion method, usable crystals were obtained and micro-seeding techniques were applied to improve the quality of crystals. The crystals used for data collection grew into a rock shape 7 days after micro-seeding (Figure 2). And the crystals of ERAP1_R-IINFEKL complex share a similar crystal shape and crystallization conditions with that of ERAP1_R-His6 recombinant protein. Detailed comparisons of crystallization conditions for the two constructs are summarized in Table 2.
Table 1. Antigenic peptide sequences attached to ERAP1 regulatory domain.
Table 2. Comparison of crystallization conditions for two protein constructs.
Figure 1. SDS PAGE analysis of recombinant ERAP1 regulatory domain (ERAP1_R) with antigenic peptide. Lane 1 is protein molecular marker (sizes labeled). Lane 2 is gel-filtration purified and concentrated ERAP1_R-IINFEKL protein (42 kDa).
Figure 2. Crystals of ERAP1_R-IINFEKL grown in 100 mM Tris, pH 8.5 and 16% PEG8000 using hanging drop vapor diffusion method at 277 K.
Reflections beyond 2.8 Å resolution were observed on diffraction images collected at the National Synchrotron Light Source―Brookhaven (Figure 3). A dataset was collected and processed to P21 space group with cell dimension a = 64.2, b = 66.8, c = 66.3 Å, β = 110.2˚. The diffraction images were processed using imosflm with an overall Rmerge of 12.5%. The detailed data collection and processing statistics are summarized in Table 3. The comparison of ERAP1_R-IINFEKL and ERAP1_R-His6 indicates that they share the same space group and have high similarity in unit cell dimension (Table 4).
Determination of the complex structure is underway by molecular replacement in CCP4 suite using ERAP1_R-His6 as a searching model (PDB entry 3RJO). To avoid model bias, coordinates of the His6 tag were removed in order to calculate a Refmac-refined omit map, which shows clear electron density for the antigenic peptides C-terminus (IINFEKL) in close contact with the ERAP1_R domain.
4. Conclusion
Multiple single-chain bimodular constructs have been designed to study proteolytic mechanism of ERAP1. Recombinant proteins of these bimodular constructs have been purified and crystallized by hanging drop vapor
Figure 3. Diffraction image of ERAP1_R-IINFEKL collected on beamline X29 at the National Synchrotron Light Source―Brookhaven. The red circle indicates the resolution of 2.8 Å.
Table 3. Data collection and processing statistics.
aNumbers in parentheses refer to the outermost (highest) resolution shell. b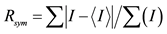 , where I is the observed intensity and
, where I is the observed intensity and 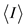 is the weighted mean of the reflection intensity.
is the weighted mean of the reflection intensity.
Table 4. Comparison of two crystal forms.
diffusion method. These crystals belong to space group P21, with unit cell parameters a =64.2, b = 66.8, c = 66.3 Å, β = 110.2˚. In the crystals, the ERAP1 regulatory domain forms an intermolecular complex with the engineered antigenic peptide. These single-chain bimodular constructs will thus facilitate high-resolution crystallographic studies on sequence-specific interactions between the ERAP1 regulatory domain and antigen peptide’s C-terminal ends.
Acknowledgements
We thank Dr. Howard Robinson for assistance on data collection, Lei Zhang and Monyrath Chan for assistance on discussions and exchange of ideas, and Ms. Victoria Calcagno for critical reading of the manuscript. This work was supported by Grants AI068831 and AI078134 from the NIH.
Cite this paper
LufeiSui,AmitGandhi,Hwai-ChenGuo, (2015) Single-Chain Expression and Crystallization of an Antigenic C-Terminus in Complex with the Regulatory Domain of ER Aminopeptidase 1. Crystal Structure Theory and Applications,04,47-52. doi: 10.4236/csta.2015.44006
References
- 1. Saric, T., Chang, S.C., Hattori, A., York, I.A., Markant, S., Rock, K.L. and Goldberg, A.L. (2002) An IFN-γ—Induced Aminopeptidase in the ER, ERAP1, Trims Precursors to MHC Class I—Presented Peptides. Nature Immunology, 3, 1169-1176. http://dx.doi.org/10.1038/ni859
- 2. York, I.A., Chang, S.C., Saric, T., Keys, J.A., Favreau, J.M., Goldberg, A.L. and Rock, K.L. (2002) The ER Aminopeptidase ERAP1 Enhances or Limits Antigen Presentation by Trimming Epitopes to 8-9 Residues. Nature Immunology, 3, 1177-1184. http://dx.doi.org/10.1038/ni860
- 3. Chang, S.C., Momburg, F., Bhutani, N. and Goldberg, A.L. (2005) The ER Aminopeptidase, ERAP1, Trims Precursors to Lengths of MHC Class I Peptides by a “Molecular Ruler” Mechanism. Proceedings of the National Academy of Sciences of the United States of America, 102, 17107-17112. http://dx.doi.org/10.1073/pnas.0500721102
- 4. Guo, H.C., Jardetzky, T.S., Garrettt, T.P., Lane, W.S., Strominger, J.L. and Wiley, D.C. (1992) Different Length Peptides Bind to HLA-Aw68 Similarly at Their Ends But Bulge out in the Middle. Nature, 360, 364-366. http://dx.doi.org/10.1038/360364a0
- 5. Fremont, D.H., Matsumura, M., Stura, E.A., Peterson, P.A. and Wilson, I.A. (1992) Crystal Structures of Two Viral Peptides in Complex with Murine MHC Class I H-2Kb. Science, 257, 919-927. http://dx.doi.org/10.1126/science.1323877
- 6. Kochan, G., Krojer, T., Harvey, D., Fischer, R., Chen, L., Vollmar, M., and Oppermann, U. (2011) Crystal Structures of the Endoplasmic Reticulum Aminopeptidase-1 (ERAP1) Reveal the Molecular Basis for N-Terminal Peptide Trimming. Proceedings of the National Academy of Sciences of the United States of America, 108, 7745-7750. http://dx.doi.org/10.1073/pnas.1101262108
- 7. Evnouchidou, I., Momburg, F., Papakyriakou, A., Chroni, A., Leondiadis, L., Chang, S.C., and Stratikos, E. (2008) The Internal Sequence of the Peptide-Substrate Determines Its N-Terminus Trimming by ERAP1. PLoS ONE, 3, e3658-e3658. http://dx.doi.org/10.1371/journal.pone.0003658
- 8. Nguyen, T.T., Chang, S.C., Evnouchidou, I., York, I.A., Zikos, C., Rock, K.L. and Stern, L.J. (2011) Structural Basis for Antigenic Peptide Precursor Processing by the Endoplasmic Reticulum Aminopeptidase ERAP1. Nature Structural & Molecular Biology, 18, 604-613. http://dx.doi.org/10.1038/nsmb.2021
- 9. Gandhi, A., Lakshminarasimhan, D., Sun, Y. and Guo, H.C. (2011) Structural Insights into the Molecular Ruler Mechanism of the Endoplasmic Reticulum Aminopeptidase ERAP1. Scientific Reports, 1, 1-5. http://dx.doi.org/10.1038/srep00186
- 10. R?tzschke, O., Falk, K., Stevanovic, S., Gnau, V., Jung, G. and Rammensee, H.G. (1994) Dominant Aromatic/Ali- phatic C-Terminal Anchor in HLA-B* 2702 and B* 2705 Peptide Motifs. Immunogenetics, 39, 74-77. http://dx.doi.org/10.1007/BF00171803�
- 11. Fleischhauer, K., Avila, D., Vilbois, F., Traversari, C., Bordignon, C. and Wallny, H.J. (1994) Characterization of Natural Peptide Ligands for HLA-B* 4402 and -B* 4403: Implications for Peptide Involvement in Allorecognition of a Single Amino Acid Change in the HLA-B44 Heavy Chain. Tissue Antigens, 44, 311-317. http://dx.doi.org/10.1111/j.1399-0039.1994.tb02401.x
NOTES

*Corresponding author.





