Open Journal of Veterinary Medicine
Vol.06 No.05(2016), Article ID:66707,10 pages
10.4236/ojvm.2016.65009
Detection of Babesia bovis and Babesia bigemina in Water Buffaloes (Bubalus bubalis) in Endemic Areas of São Paulo State, Brazil
T. A. Néo1, R. Giglioti2*, D. Obregón3, T. B. Bilhassi2, H. N. Oliveira2, R. Z. Machado4, F. de F. Aníbal1, L. G. Brito5, W. Malagó Jr.6, F. A. Bressani6, M. C. S. Oliveira6
1Department of Morphology and Pathology, Universidade Federal de São Carlos, São Carlos, Brazil
2Department of Animal Science, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista, Jaboticabal, Brazil
3Agrarian University of Havana, San José de las Lajas, Cuba
4Department of Veterinary Pathology, Faculdade de Ciências Agrárias e Veterinárias, Universidade Estadual Paulista, Jaboticabal, Brazil
5Embrapa Rondônia, Porto Velho, Brazil
6Embrapa Pecuária Sudeste, São Carlos, Brazil

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 9 April 2016; accepted 20 May 2016; published 24 May 2016
ABSTRACT
Babesiosis is a tick-transmitted disease that causes severe economic losses to the cattle industry in Brazil. Water buffaloes (Bubalus bubalis) are often carriers of Babesia spp., but there are no studies that provide an accurate estimation of this infection in animals raised in regions of endemic stability. This study was conducted to investigate Babesia bovis and B. bigemina infections in 108 water buffaloes (50 calves and 58 adult females) located in areas of São Paulo state, where the animals were continuously exposed to Rhipicephalus microplus ticks. B. bovis and B. bigemina infections were screened by microscopic examination of blood smears, nested PCR (nPCR) and quantitative real-time PCR (qPCR), which were also used to estimate the number of copies (NC) of the cytochrome b (mt-cytB) gene in the blood samples. B. bigemina was found in blood smears of three calves from Alambari herd (all with less than 0.1% parasitemia). Molecular techniques were more sensitive than blood smears to diagnose piroplasms in water buffaloes: 20.37% and 100.00% for B. bovis-infected animals and 59.26% and 100.00% for B. bigemina-infected animals, respectively for nPCR and qPCR. The NC of mt-cytB gene of B. bovis and B. bigemina in blood samples revealed significant effects (p < 0.05) of herd-age, species and their interaction. The NC values were higher (p ≤ 0.05) for B. bigemina (2.80 ± 0.06) than for B. bovis (2.61 ± 0.05). Within each herd-age, differences between the species’ NC values were found only in Alambari calves, which showed significantly higher (p ≤ 0.05) NC of B. bigemina (3.48 ± 0.13). The calves and cows from Ibaté showed the lowest NC of B. bigemina (2.29 ± 0.13 and 2.63 ± 0.14) and B. bovis (2.54 ± 0.11 and 2.37 ± 0.12), respectively. These data suggest a high prevalence of B. bovis and B. bigemina infection in the buffalo population in endemic areas of São Paulo state.
Keywords:
Bubalus bubalis, Babesiosis, Diagnosis, nPCR, qPCR

1. Introduction
Bovine babesiosis is a tick-borne disease that occurs worldwide, causing significant losses to livestock breeders [1] - [3] . In most of Brazil, endemic stability for Babesia bovis and B. bigemina is common [4] [5] . After primary infection, recovered animals frequently sustain subclinical infections and may continue to infect the tick vectors [2] [6] [7] . In acute babesiosis, the infection can be diagnosed by the identification of piroplasms in stained blood smears using microscopic examination, but in parasite carriers, the detection requires more sensitive methods [8] - [10] . Nested PCR (nPCR) has high sensitivity and specificity [9] , whereas quantitative real-time PCR (qPCR) combines the detection of target template with quantification [11] , having the advantages of reduced time and lower risk of cross-contamination [12] .
In 2001, there were some three million water buffaloes in Brazil, with a population growth rate estimated at 10% a year [13] . In the country’s southeastern region, buffalo milk is mainly used to produce mozzarella cheese. The growing population of these animals is largely due to the high aggregate value of this product. Previous studies in tropical regions have indicated that water buffaloes are often carriers of Babesia spp. due to common infestation by the cattle tick Rhipicephalus microplus [14] [15] . However, there are no studies providing an accurate estimation of Babesia spp. infection in water buffaloes in a region of endemic stability. The increasing introduction of water buffalo dairy herds in the state of São Paulo led us to study the extent of infection in these animals using techniques considered as having high sensitivity and specificity. Different diagnostic methods were employed and compared to assess their suitability.
2. Materials and Methods
2.1. Animals and Sample Collection
The experimental group consisted of 108 clinically healthy water buffaloes (58 adult females aged between 3 - 5 years old and 50 calves aged 1 - 3 months old) from four municipalities in São Paulo, in southeastern Brazil. All animals were sampled only once, from January to February 2014, in four dairy herds: São Carlos (22˚01' S and 47˚53' W; n = 14 adult females); Ibaté (21˚57' S and 47˚59' W; n = 48, 25 calves and 23 adult females); Alambari (23˚33' S and 47˚53' W; n = 25 calves); and Dourado (22˚06' S and 48˚19' W; n = 21 adult females). None of the sampled herds was subjected to tick control. The blood samples were collected from the caudal vein using EDTA Vacutainers® (Becton Dickinson) for DNA extraction and packed cell volume (PCV) determination by the microhematocrit method [16] . Blood samples from ear vessels were used to prepare thin blood smears for determination of parasitemia (%). Total genomic DNA was extracted from 300 µl of each blood sample with the Easy DNATM kit (Invitrogen, USA), as recommended by the manufacturer. DNA samples were eluted in 100 µl of Tris-EDTA buffer and maintained in a freezer at −80˚C.
This experiment was conducted according to the ethical principles of animal experimentation of Embrapa Southeast Livestock ethics committee for animal experiments (CEUA PRT 05/2014).
2.2. nPCR Procedures
Previously described protocols were used for the nPCR assays for detection by specific amplification of DNA fragments of B. bovis [17] and B. bigemina [18] . Assays were performed in a volume of 25 μl: 12.5 μl of Master Mix red (Ampliqon, Denmark), 5.5 μl of ultrapure water (Invitrogen, USA), 10 pm of each primer (Table 1) and 5 μl of template DNA. The same buffer and 2 μl of PCR reactionmedium were used as the nPCR template. Optimal primer annealing temperatures were determined experimentally for all PCR protocols in a Mastercycler Gradient thermocycler (Eppendorf). The optimal PCR thermocycling conditions for B. bovis PCR were: initial denaturation at 95˚C for 1 min; 35 cycles: at 95˚C for 1 min (denaturation); 60˚C for 1 min (annealing); 72˚C (extension) for 1 min; and a final extension at 72˚C for 5 min. The nPCR assays were performed under the same PCR conditions, except the annealing step, which was at 68˚C. The conditions for B. bigemina PCR amplifications were: initial denaturation at 95˚C for 2 min; 40 cycles of 94˚C for 1 min (denaturation), 66˚C for 1 min (annealing), 72˚C for 1 min (extension) and a final extension at 72˚C for 5 min. The nPCR conditions were identical except the annealing step, which was at 69˚C for 30 s. Purified samples of B. bovis and B. bigemina (provided by the Department of Pathology, School of Veterinary Medicine, São Paulo State University, Jaboticabal) were submitted to DNA extraction and used as positivecontrol. DNA samples were considered positive if in nPCR specific bands were visualized on 2% agarose gel (B. bovis = 290 bp and B. bigemina = 262 bp). To further confirm the nPCR results, positive samples were selected from each herd and were cloned into the pGEMÒ-T Easy Vector System (Promega, Madison, USA). Recombinant clones were used to transform Escherichia coli DH5α-competent cells. Plasmids containing the insert were purified using a QIAprep spin miniprep kit (Qiagen), and the amplicons were sequenced in an Applied Biosystems ABI Prism® 3100 Avant genetic analyzer (Foster City, CA). The obtained sequences were deposited in the GenBank and subjected to BLAST analysis [19] .
2.3. qPCR Procedures
The qPCR tests were performed using the CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad), with the primers that amplify an 88-bp fragment from the mt-cytB gene of each Babesia species [20] . Genomic DNA extracted from isolates of B. bovis and B. bigemina were used to construct the calibration curve and as positive control.
The optimized qPCR was prepared in a volume of 15 μl, with 7.5 μl of Eva Green® Supermix (Bio-Rad), 0.12 μM of each primer, cbisg-1 and cbisg-2 for B. bigemina and cbosg-1 and cbosg-2 for Babesia bovis (Table 1), 5.26 μl of ultrapure water (Invitrogen, USA) and 2.0 μl of template DNA. The thermocycling conditions were: 2 minutes at 95˚C, 40 cycles at 95˚C for 15 s and annealing and extension at 57˚C for one min for B. bovis
Table 1. Primer sets used for nPCR and qPCR analysis of Babesia bovis and Babesia bigemina.
*Pb identified in Nucleotide BLAST (Sequences ID: gb|KU714606.1| and gb|KU714605.1|).
and B. bigemina. The qPCR reactions were carried out in duplicate, and each run included ddH20 (Invitrogen, USA) as a negative control, DNA from piroplasms isolates and a serial dilution of plasmid DNA. To prevent contamination, the reactions were prepared using tips with barriers, in rooms specifically isolated for this purpose.
To estimate the NC of the mt-cytB gene of B. bovis and B. bigemina in blood samples, calibration curves were standardized from the cloned products from the B. bovis and B. bigemina isolates. qPCR amplification products of the two parasites were purified with the PureLinkTM PCR purification kit (Invitrogen), connected to a pGEMÒ-T Easy Vector System (Promega, Madison, USA) for subsequent transformation of the E. coli DH5α- competent cells. The white colonies were selected, amplified to confirm the vector insert and then grown in SOB medium at 37˚C ± 1˚C overnight under shaking. The DNA was extracted using the PureLink® Quick Plasmid Miniprepkit (Invitrogen) and sequenced with an Applied Biosystems ABI Prism 3130 AvantÒ genetic analyzer. The sequences obtained were submitted to BLAST analysis [19] . Plasmid concentration was quantified using a NanoDrop ND-1000 spectrophotometer, and serially diluted (10−1 to 10−10) to determine the limits of quantification of each species. Each dilution was subjected to qPCR assays with the samples and controls to estimate the NC of mt-cytB gene of B. bovis and B. bigemina [21] . The NC values found in the dilutions from the calibration curves were used to establish a regression equation allowing estimation of the NC of mt-cytB gene values for the DNA from B. bovis and B. bigemina for each sample. All amplifications and data were analyzed with the CFX96 software (BioRad). The efficiency (E) of reaction, expressed in percentage, was determined by the following formula: 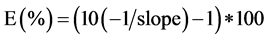 [22] [23] .
[22] [23] .
qPCR analytical sensitivity was estimated from purified products of B. bovis and B. bigemina isolates in a final DNA concentration of 1 ng/ul. The samples were submitted to 10-fold serial dilutions with five repetitions. The NC of the mt-cytB gene in each dilution was calculated [21] . The cutoff Cq (quantification cycle) was established for each sample according to the result of qPCR sensitivity tests and corresponded to the last serial dilution where more than 50% of the five replicates were amplified [24] .
2.4. Statistical Analysis
The PCV values were subjected to one-way analysis of variance to check the combined effect of herd-age, using the SAS®-GLM procedure. For each Babesia species, Fisher’s exact test was applied to verify whether the frequencies of positive animals detected by the nPCR assays were similar between the different herd-age variables. The NC data, transformed into log10 (NC + 1) for both Babesia species, were analyzed together using the SAS MIXED procedure. In this analysis, the measures of NC of B. bovis and B. bigemina were considered as repeated measures of the same variable. So, the model of analysis included the effect of Babesia species (B. bovis and B. bigemina) and the combined effect of herd-age along with the interaction between them as fixed effects and the effect of the animal as a random effect. An unstructured (co)variance matrix was assumed for the residuals to account for possible variance heterogeneity between the two species. This model permits estimating the correlation between the infection rates of the two Babesia species. Means of NC and PCV were compared by the Tukey test and considered statistically significant if p ≤ 0.05. The Statistical Analysis System v.9.1 [25] was used in all the analyses.
3. Results
3.1. Blood Analysis
The microscopic examination of the blood smears revealed positive samples only for B. bigemina infected calves from Alambari (3/25), all with parasitemia below 0.1%.
The mean PCV (Table 2) values obtained for the adult females from all the herds and the calves from Alambari were similar (no significant difference), while the mean for the calves from Ibaté were significantly higher (p < 0.05).
3.2. nPCR and qPCR
The amplicons sequenced from the rap1 gene of B. bovis (KC907706, KC907707, KC907704, KC907705, KP843858) and 18S ribosomal RNA gene of B. bigemina (KF153076, KC333880.1 KC858975, KC858976.1, KR526274) showed high similarity (98% - 100% for B. bovis and 93% - 100% for B. bigemina) with those already deposited in the GenBank. Representative gel electrophoresis results for the B. bovis and B. bigemina PCR and nPCR are showed in Figure 1.
The nPCR assays detected 13.5% infection by B. bigemina of calves (27/50, including the three calves that were positive by microscopic detection) and 63.7% of adult females (37/58). In turn, 11 animals were positive for B. bovis in each category, representing 22% and 18.9% of the calves and adult females examined, respectively. All told, nPCR detected 64animals infected by B. bigemina and 22 by B. bovis. Fisher’s exact test detected variation in the infection rates between herd-age for both species (p < 0.05). As can be seen in Table 3, the herd-age group with highest infection frequency by both B. bovis and B. Bigemina was the calves from Alambari, while the calves from Ibaté presented the lowest positive rate by nPCR, also for both species. For the adult females, the lowest infection rate by B. bigemina was observed in the herd from São Carlos, while for B. bovis the frequencies were similar among the adult animals in all three herds.
The qPCR assays showed 100% of the animals were infected, irrespective of category or origin (Table 3). The qPCR cutoff Cq was established at 39 cycles for the two piroplams, corresponding to dilutions with 20 copies of mt-cytB gene/µl, which were the lowest concentrations at which three of the five repetitions were amplified (≥50%). All the samples were classified as qPCR positive, based on the melting peaks and Cq (before Cq 39, Figure 2). The positive samples showed melting temperatures of 76.0˚C ± 0.26˚C for B. bigemina and 77.5˚C ± 0.24˚C for B. bovis (Figure 2). The amplified sequences of the mt-cytB gene were identical (100% similarity) to those of previous samples deposited in the GenBank: AB 499088.1 and GQ 214235.1 for B. bovis and LK 054939.1 and AB 499085.1 for B. bigemina.
Table 2. Least square means and standard errors of the packed cell volume (PCV, %) in water buffaloes according to age and herd.
Means followed by the different letters in columns differ significantly (p < 0.05).
Figure 1. Agarose gel electrophoresis of B. bovis (A) and B. bigemina (B) PCR and nested-PCR. A: line 1 = standard base pairs; lines 2 - 4 = B. bovis PCR positive samples; lines 5 - 7 = B. bovis nested-PCR positive samples; line 8 = negative control. B: line 1 = standard base pairs; lines 2 - 4 = B. bigemina PCR positive samples; lines 5 - 7 = B. bigemina nested-PCR positive samples; line 8 = negative control.
Table 3. Summary of the molecular detection of Babesia bovis and Babesia bigemina in water buffaloes determined by nPCR and qPCR, according to age and herd.
Figure 2. (A) Amplification of the fragment of the mt-cytB gene of B. bovis; (B) amplification of the fragment of the mt-cytB gene of B. bigemina; (C) melt peak derived from amplification of the fragment of the mt-cytB gene of B. bovis; and (D) melt peak derived from amplification of the fragment of the mt-cytB gene of B. bigemina.
Statistical analysis of the NC of the mt-cyB gene of B. bovis and B. bigemina showed significant effects (p ≤ 0.05) of herd-age, species, and the interactions. The NC values were higher (p ≤ 0.05) for B. bigemina (2.80 ± 0.06) than for B. bovis (2.61 ± 0.05). Within each herd-age, differences between the species’ NC values were found only in Alambari calves, which showed significantly higher (p ≤ 0.05) NC of B. bigemina (3.48 ± 0.13). The calves and cows from Ibaté showed the lowest NC for B. bigemina (2.29 ± 0.13 and 2.63 ± 0.14) and B. bovis (2.54 ± 0.11 and 2.37 ± 0.12), respectively (Table 4).
Table 4. Least-square means and standard errors of the number of Mt-cytB gene copies (NC) of Babesia bovis and Babesia bigemina by qPCR in water buffalo blood samples, according to age and herd.
*Means followed by the different capital letters in the rows differ significantly (p < 0.05); **Means followed by the same small letters in the columns do not differ significantly (p > 0.05).
4. Discussion
In this experiment, we found a high frequency of infection by B. bovis and B. bigemina in apparently healthy buffaloes raised in São Paulo state, using PCR-based tests. Blood samples from animals with different ages were submitted to varied method to diagnose infection by hemoparasites: direct examination, nPCR and qPCR. The PCV data were used as indicators of animal health.
All the herds studied coexist with R. microplus, a tick that is widely distributed in Brazil [26] . The detection of calves with patent parasitemia by B. bigemina and the high frequency of positive cows and calves for infection by both Babesia species in the qPCR assays were similar to the patter observed for cattle raised in the same region, characterized as having stable endemia [7] [27] - [29] .
The average PCV values of the animals were comparable to those described for healthy cattle [16] , which can vary from 24% to 46%. A study in Italy revealed that buffalo calves had higher PCV levels, with gradual decline as the animals got older [30] . In Brazil, some authors have also found a similar pattern to that observed in Italy, i.e., the PCV declined with the age of the buffaloes [31] - [34] . In this study, we observed that the calves from Alambari presented PCV levels similar to those of the adult animals, and significantly lower than those observed in the calves from Ibaté. The average PCV levels found for the calves from Alambari and Ibaté in this study (34.60 ± 1.20 and 46.40 ± 1.20) differed from that found [33] in the same region of Brazil, which was 40.6 ± 3.2. The elimination of the fetal erythrocytes can cause significant reductions in the levels of these blood cells in animals with ages between 3 and 4 months old [16] , which might be the cause of the lower PCV levels found in the calves from Alambari. Besides this, it has been observed that buffaloes can present variations in the PCV levels, depending on age, breeding region, nutritional and sanitary management [35] [36] . This can explain, at least in part, the differences observed in the average PCV levels of the calves from Alambari and Ibaté.
The direct examination of blood smears compared with nPCR assays confirmed that the sensitivity of the first test was not sufficient to detect B. bovis and B. bigemina in carrier animals, as also observed by other researchers [8] [9] . PCR reactions can detect DNA quantities on the order of 1 pg and this sensitivity can be increased to 1 fg by using nPCR [37] [38] , with the further advantage that the limits of specificity can be defined according to the particular needs of each diagnosis [39] . Despite, this, false negative results can occur, due to the presence of substances that inhibit polymerase that can be co-purified with the DNA [40] , because the sequence identified by the primers is absent, inaccessible or only present in very small quantities [41] .
The results of the qPCR assays allowed detecting a greater number of positive animals than those of nPCR, with some advantages, such as speed, robustness, precision and low contamination risk, in addition to the possibility of quantification [42] . Buling et al. [20] compared the performance of the qPCR assay using the mt-cytB gene as the target (also used in our experiment) and conventional PCR employing the 18s rRNA gene from B. bovis and B. bigemina in DNA samples extracted from the blood of 80 horses. The authors detected one horse infected by B. bovis and two by B. bigemina in the 80 blood samples analyzed by qPCR, but none were positive by the standard ribosomal PCR [20] . Ramos et al. detected a larger number of positive animals for B. bovis when using a qPCR protocol (msa2c gene) than when using conventional PCR for the mt-cytB gene [43] . Despite the high sensitivity of the qPCR technique, it also has drawbacks, such as the need for previous knowledge of the target gene [11] and the impossibility of discriminating the amplicons by size [41] .
The distribution of infected animals in the herds studied here showed that buffaloes in the state of São Paulo were carriers of B. bovis and B. bigemina, as has been observed in other regions of Brazil and other tropical countries where tick-borne diseases were endemic [11] [14] [15] [44] . The results obtained with qPCR show that in stable endemic areas, infection by both Babesia species is very common in water buffaloes.
Regarding the NC hemoparasite DNA in buffaloes, we did not find any data in the literature. Bilhassi et al. [45] found higher average NC values of mt-cytB for B. bovis in both cows and calves of different cattle breeds, 5.00 ± 0.07 and 4.41 ± 0.07 respectively, compared with our results for adults (from 2.37 ± 0.12 to 2.81 ± 0.15) and calves (2.54 ± 0.11 and 2.78 ± 0.11). The mechanisms that assure low parasitemia of B. bovis and B. bigemina in water buffalo hosts are unknown, although it is know that babesias persistently infect various vertebrates without causing any symptoms [46] [47] .
In this experiment, the use of qPCR allowed finding high frequencies of infection by B. bovis and B. bigemina in both buffalo calves and cows raised in endemic areas of the state of São Paulo. Further studies should be conducted to clarify the real impact of these infections on buffalo herds.
Acknowledgements
This project was supported by funding from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-474648/210-9) and the Brazilian Agricultural Research Corporation (Embrapa). CAPES, NCPq, TWAS and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) provided research grants to the authors.
Conflict of Interest Statement
None of the authors of this study have a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
Cite this paper
T. A. Néo,R. Giglioti,D. Obregón,T. B. Bilhassi,H. N. Oliveira,R. Z. Machado,F. de F. Aníbal,L. G. Brito,W. Malagó Jr.,F. A. Bressani,M. C. S. Oliveira, (2016) Detection of Babesia bovis and Babesia bigemina in Water Buffaloes (Bubalus bubalis) in Endemic Areas of São Paulo State, Brazil. Open Journal of Veterinary Medicine,06,75-84. doi: 10.4236/ojvm.2016.65009
References
- 1. McCosker, P.J. (1981) The Global Importance of Babesiosis. In: Ristic, M. and Kreier, J.P., Eds., Babesiosis, Academic Press, New York.
- 2. Bock, R., Jackson, L., De Vos, A. and Jorgensen, W. (2004) Babesiosis of Cattle. Parasitology, 129, S247-S269.
- 3. Uilenberg, G. (2006) Babesia—A Historical Overview. Veterinary Parasitology, 138, 3-10.
http://dx.doi.org/10.1016/j.vetpar.2006.01.035 - 4. Kessler, R.H., Madruga, C.R., Schenk, M.A. and Ribeiro, O.C. (1983) Babesiose cerebral por Babesia bovis em bezerros no Estado do Mato Grosso do Sul. Pesquisa Agropecuária Brasileira, 18, 931-935.
- 5. Vidotto, O., Andrade, G.M., Amaral, C.H.S., Barbosa, C.S., Freire, R.L., Rocha, M.A. and Vidotto, M.C. (1997) Frequência de anticorpos contra Babesia bigemina, Babesia bovis e Anaplasma marginale em rebanhos leiteiros da região de Londrina, Paraná. Arquivo Brasileiro de Medicina Veterinária e Zootecnia, 49, 655-659.
- 6. Calder, J.A.M., Reddy, G.R., Chieves, L., Courtney, C.H., Littell, R., Livengood, J.R., Norval, R.A.I., Smith, G. and Dame, J.B. (1996) Monitoring Babesia bovis in Cattle Using PCR-Based Tests. Journal of Clinical Microbiology, 34, 2748-2755.
- 7. Oliveira-Sequeira, T.C.G., Oliveira, M.C.S., Araujo-Júnior, J.P. and Amarante, A.F.T. (2005) PCR-Based Detection of Babesia bovis and Babesia bigemina in Their Natural Host Boophilus microplus and Cattle. International Journal for Parasitology, 35, 105-111.
http://dx.doi.org/10.1016/j.ijpara.2004.09.002 - 8. Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A. and Speleman, F. (2002) Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biology, 3, RESEARCH0034.
- 9. Caraguel, C.G.B., Stryhn, H., Gagne, N., Dohoo, I. and Hammell, K.L. (2011) Selection of a Cutoff Value for Real-Time Polymerase Chain Reaction Results to Fit a Diagnostic Purpose: Analytical and Epidemiologic Approaches. Journal of Veterinary Diagnostic Investigation, 23, 2-15.
http://dx.doi.org/10.1177/104063871102300102 - 10. SAS Institute (2002/2003) SAS/Insight User’s Guide. Version 9.1.3 for Windows. SAS Institute, Cary.
- 11. Horn, S.C. and Arteche, C.C.P. (1984) Carrapato, Berne e Bicheira no Brasil, 1983. Secretaria de Defesa Sanitária Animal do Ministério da Agricultura, Brasília, 153 p.
- 12. Gonçalves, P.M. (2000) Epidemiology and Control of Bovine Babesiosis and Anaplasmosis in the Southeast Region of Brazil. Ciência Rural, 30, 187-194.
- 13. Oliveira, M.C.S., Oliveira-Sequeira, T.C.G., Araujo-Júnior, J.P., Amarante, A.F.T. and Oliveira, H.N. (2005) Babesia spp. Infection in Boophilus microplus Engorged Female and Eggs in São Paulo State, Brazil. Veterinary Parasitology, 130, 61-67.
http://dx.doi.org/10.1016/j.vetpar.2005.03.007 - 14. Oliveira, M.C.S., Oliveira-Sequeira, T.C.G., Regitano, L.C.A.; Alencar, M.M., Néo, T.A., Silva, A.M. and Oliveira, H.N. (2008) Detection of Babesia bigemina in Cattle of Different Genetic Groups and in Rhipicephalus (Boophilus) microplus Tick. Veterinary Parasitology, 155, 281-286.
http://dx.doi.org/10.1016/j.vetpar.2008.04.022 - 15. Ciaramella, P., Corona, M., Ambrosio, R., Consalvo, F. and Persechino, A. (2005) Haematological Profile on Non-Lactating Mediterranean Buffaloes (Bubalus bubalis) Ranging in Age from 24 Months to 14 Years. Research in Veterinary Science, 79, 77-80.
http://dx.doi.org/10.1016/j.rvsc.2004.11.004 - 16. Silva, M.B., D’Angelino, J.L., Araujo, W.P., Galhardo, M., Garcia, M. and Birgel, F.H. (1992) Avaliação do eritrograma de búfalos (Bubalus bubalis) criados na região do Vale do Ribeira em São Paulo. Brazilian Journal of Veterinary Research and Animal Science, 29, 113-119.
- 17. Costa, C.L., Kohayagawa, A., Dell Porto, A. and Bonfim, S.E.M. (1997) Determinação dos níveis de anticorpos anti-Babesia spp. em bezerros bubalinos (Bubalus bubalis), desde o nascimento até um ano de. idade. Revista brasileira de parasitologia veterinaria, 6, 117-121.
- 18. Gomes, V., Moura, J.A., Madureira, K.M., Baptistella, F., Kitamura, S.S., Blagitz, M.G. and Benesi, F.J. (2010) Valores de referência e influência da idade no eritrograma de bubalinos da raça Murrah. Pesquisa Veterinária Brasileira, 30, 301-304.
- 19. França, R.T., Lopes, S.T.A., Martins, D.B., Costa, M.M., Leal, M.L.R., Mazzanti, C.M.A., Schuh, R. and Dornelles, G.L. (2011) Valores hematológicos de búfalos em diferentes faixas etárias criados na região central do Rio Grande do Sul. Revista Brasileira de Ciência Veterinária (Impresso), 18, 51-54.
- 20. Ferrer, J.M., árraga, C.M. and Barboza, M. (2000) Caracterización hematológica de laespecieBubalus bubalis por sexo y edad. Revista Científica FCV-LUZ, 10, 508-514.
- 21. Fernández, A.H., Romero, O., Montiel, N., Trujillo, H.N. and Cahuao, N. (2005) Determinación de valores de referenciahematologicosen búfalas (Bubalusbubalis) preparto e postpartoen una unidad de producciónenelsurdel lago de Maracaíbo, Venezuela. Revista Científica FCV-LUZ, 15, 119-124.
- 22. Hayden, J.D., Ho, S.A., Hawkey, P.M., Taylor, G.R. and Quirke, P. (1991) The Promises and Pitfalls of PCR. Reviews in Medical Microbiology, 2, 129-137.
- 23. Böse, R., Jorgensen, W.K., Dalgliesh, R.J., Friedhoff, K.T. and de Vos, A.J. (1995) Current State and Future Trends in the Diagnosis of Babesiosis. Veterinary Parasitology, 57, 61-74.
http://dx.doi.org/10.1016/0304-4017(94)03111-9 - 24. Barker Jr., R.H. (1990) DNA Probe Diagnosis of Parasitic Infections. Experimental Parasitology, 70, 494-499.
http://dx.doi.org/10.1016/0014-4894(90)90136-z - 25. Hunt, P.W. (2011) Molecular Diagnosis of Infections and Resistance in Veterinary and Human Parasites. Veterinary Parasitology, 180, 12-46.
http://dx.doi.org/10.1016/j.vetpar.2011.05.027 - 26. Barker, R.H., Banchongaksorn, T., Couval, J.M., Suwonkerd, W., Rimwungtragoon, K. and Wirth, D.F. (1994) Plasmodium falciparum and P. Vivax: Factors Affecting Sensitivity and Specificity of PCR-Based Diagnosis of Malaria. Experimental Parasitology, 79, 41-49.
http://dx.doi.org/10.1006/expr.1994.1057 - 27. Bastien, P., Procop, G.W. and Reischl, U. (2008) Quantitative Real-Time PCR is Not More Sensitive than “Conventional” PCR. Journal of Clinical Microbiology, 46, 1897-1900.
http://dx.doi.org/10.1128/jcm.02258-07 - 28. Ramos, C.A.N., Araújo, F.R., Souza, I.I.F., Bacanelli, G., Luiz, H.L., Russi, L., Oliveira, R.H.M., Soares, C.O., Rosinha, G.M.S. and Alves, L.C. (2011) Real-Time Polymerase Chain Reaction Based on msa2c Gene for Detection of Babesia bovis. Veterinary Parasitology, 176, 79-83.
http://dx.doi.org/10.1016/j.vetpar.2010.10.035 - 29. Mahmmod, Y. (2014) Natural Babesia bovis Infection in Water Buffaloes (Bubalus bubalis) and Crossbred Cattle under Field Conditions in Egypt: A Preliminary Study. Journal of Arthropod-Borne Diseases, 8, 1-9.
- 30. Bilhassi, T.B., Oliveira, H.N., Ibelli, A.M.G., Giglioti, R., Regitano, L.C.A., Oliveira-Sequeira, T.C.G., Bressani, F.A., Malagó Jr., W., Resende, F.D. and Oliveira, M.C.S. (2014) Quantitative Study of Babesia bovis Infection in Beef Cattle from São Paulo state, Brazil. Ticks and Tick-Borne Diseases, 5, 234-238.
http://dx.doi.org/10.1016/j.ttbdis.2013.11.002 - 31. Homer, M.J., Aguilar-Delfin, I., Telford, S.R., Krause, P.J. and Persing, D.H. (2000) Babesiosis. Clinical Microbiology Reviews, 13, 451-469.
- 32. Chauvin, A., Moreau, E., Bonnet, S., Plantard, O. and Malandrin, L. (2009) Babesia and Its Hosts: Adaptation to Long-Lasting Interactions as a Way to Achieve Efficient Transmission. Veterinary Research, 40, 37.
http://dx.doi.org/10.1051/vetres/2009020 - 33. Pfaffl, M.W. (2001) A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Research, 29, e45.
http://dx.doi.org/10.1093/nar/29.9.e45 - 34. Ke, G.M., Cheng, H.L., Ke, L.Y., Ji, W.T., Chulu, J.L., Liao, M.H., Chang, T.J. and Liu, H.J. (2006) Development of a Quantitative Light Cycler Real-Time RT-PCR for Detection of Avian Reovirus. Journal of Virological Methods, 133, 6-13.
http://dx.doi.org/10.1016/j.jviromet.2005.09.011 - 35. Buling, A., Criado-Fornelio, A., Acenzo, G., Benitez, D., Barba-Carretero, J.C. and Florin-Cristensen, M. (2007) A Quantitative PCR Assay for the Detection and Quantification of Babesia bovis and B. bigemina. Veterinary Parasitology, 147, 16-25.
http://dx.doi.org/10.1016/j.vetpar.2007.03.031 - 36. Altschul, S.F., Gish, W., Myers, E.W. and Lipman, D.J. (1990) Basic Local Alignment Search Tool. Journal of Molecular Biology, 215, 403-410.
http://dx.doi.org/10.1016/S0022-2836(05)80360-2 - 37. Guerrero, F.D., Bendele, K.G., Davey, R.B. and George, J.E. (2007) Detection of Babesia bigemina Infection in Strains of Rhipicephalus (Boophilus) microplus Collected from Outbreaks in South Texas. Veterinary Parasitology, 145, 156-163.
- 38. Figueroa, J.V., Chieves, L.P., Johnson, G.S. and Buening, G.M. (1993) Multiplex Polymerase Chain Reaction-Based Assay for the Detection of Babesia bigemina, Babesia bovis and Anaplasma marginale DNA in Bovine Blood. Veterinary Parasitology, 50, 69-81.
http://dx.doi.org/10.1016/0304-4017(93)90008-B - 39. Jain, N.C. (1993) Essentials of Veterinary Hematology. Lea &Febiger, Philadelphia, 417 p.
- 40. Silva, J.B., André, M.R., Fonseca, A.H., Lopes, C.T.A., Lima, D.H.S., Andrade, S.J.T., Oliveira, C.M.C. and Barbosa, J.D. (2013) Molecular and Serological Prevalence of Babesia bovis and Babesia bigemina in Water Buffaloes in the North Region of Brazil. Veterinary Parasitology, 197, 678-681.
- 41. Obregón, D., Oliveira, M.C.S., Tizioto, P.C., Funnes, M.E., Martínez, S., Roque, E., Fonseca, A.H. and Corona, B. (2012) Diagnóstico de Babesia bovisen búfalos de laregionoccidental de Cuba a través de unensayo de PCR. Revista de Salud Animal, 34, 101-108.
- 42. Borghese, A. and Mazzi, M. (2005) Buffalo Population and Strategies in the World. In: Buffalo Production and Research, FAO, Rome.
http://www.fao.org/docrep/010/ah847e/ah847e00.htm - 43. OIE (World Organization for Animal Health) (2012) Babesiosis Bovina. In: OIE Manual Editor, Manual de la OIE sobre animales terrestres, OIE Biological Standards Commission, Paris, 1-16.
- 44. Smith, C.J. and Osborn, A.M. (2009) Advantages and Limitations of Quantitative PCR (q-PCR)-Based Approaches in Microbial Ecology. FEMS Microbiology Ecology, 67, 6-20.
http://dx.doi.org/10.1111/j.1574-6941.2008.00629.x - 45. Terkawi, M.A., Huyen, N.X., Shinuo, C., Inpankaew, T., Maklon, K., Aboulaila, M., Ueno, A., Goo, Y., Yokoyama, N., Jittapalapong, S., Xuan, X. and Igarashi, I. (2011) Molecular and Serological Prevalence of Babesia bovis and Babesia bigemina in Water Buffaloes in the Northeast Region of Thailand. Veterinary Parasitology, 178, 201-207.
http://dx.doi.org/10.1016/j.vetpar.2011.01.041 - 46. Almeria, S., Castellà, J., Ferrer, D., Oortuño, A., Estrada-Peña, A. and Gutierrez, J.F. (2001) Bovine Piroplasms in Minorca (Balearic Islands, Spain): A Comparison of PCR-Based and Light Microscopy Detection. Veterinary Parasitology, 99, 249-259.
http://dx.doi.org/10.1016/S0304-4017(01)00464-2 - 47. Fahrimal, Y., Goff, W. and Jasmer, D.P. (1992) Detection of Babesia bovis Carrier Cattle by Using Polymerase Chain Reaction Amplification of Parasite DNA. Journal of Clinical Microbiology, 30, 1374-1379.
NOTES

*Corresponding author.




