Advances in Microbiology
Vol.05 No.07(2015), Article ID:57719,6 pages
10.4236/aim.2015.57050
Impact of Leucine 278 Residue on Fatty Acid Length Specificity of Candida antarctica Lipase B
Fanghua Wang1, Shulin Hou2, Qian Wang3, Pu Wang3, Jinsong Liu2, Bo Yang3, Yonghua Wang1*
1College of Light Industry and Food Sciences, South China University of Technology, Guangzhou, China
2State Key Laboratory of Respiratory Disease, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China
3School of Bioscience and Bioengineering, South China University of Technology, Guangzhou, China
Email: *yonghw@scut.edu.cn
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 9 June 2015; accepted 30 June 2015; published 3 July 2015
ABSTRACT
Structural analysis of Candida antarctica lipase B (CALB) indicates that side chain of leucine at 278 site lies above the entrance of the catalytic pocket, which prognosticates its potential role on substrate specificity of the enzyme. To verify this presumption, shortened side chain of glycine or proline was rational designed and mutants were constructed by site-directed mutagenesis method. The colorimetric assay using p-nitrophenyl esters of fatty acids with various chain-lengths was used to study the substrate preference of lipases. Results indicated that L278G or L278P mutations both induced the drift of substrate specificity of CALB from p-nitrophenyl caprylate (pNP-C8) to longer carbon chain length of p-nitrophenyl caprate (pNP-C10). Meanwhile, Vmax value of two mutants to pNP-C10 was both higher than that of wild-type. Docking results also indicated that shortened side chain of glycine or proline residues substitution at this site could get rid of the space block present above the catalytic pocket, and made longer chain substrate (pNP-C10) enter into the catalytic pocket easier. The modulation of specificity observed allowed for building substrate binding model and opened new possibilities for designing ligand specific lipases.
Keywords:
Candida antarctica Lipase B (CALB), Site-Directed Mutagenesis, Substrate Specificity, Stereospecific Blockade

1. Introduction
Lipase B from Candida antarctica (CALB) is a highly versatile biocatalyst mainly used for organic synthesis of the laboratory and the commercial scale. Due to the inherent activity, specificity, stability, or enantioselectivity of enzyme are not always suitable for industrial applications. Thus, engineering and modification of the natural enzyme are an important issue for protein engineers. In recent years, a number of rational and combinatorial protein engineering projects have focused on extending and tailoring catalytic and physical properties to further expand the functionality of CALB [1] . During these processes, some residue sites were screened out and had great correlation with the activity, enantioselectivity or thermo-stability of the CALB, such as Leu 278, Leu281, Leu 219, Val210, and V221 [2] - [5] . Mutations on these hotspot sites could improve the function of the CALB significantly. For the 278 site, it had been found that L278P variant showed substantially higher activity toward p-nitrophenyl palmitate (pNPP) than the wild-type CALB (WT-CALB) [2] . The kcat value of L278P mutants increased approximately 6-fold higher than WT-CALB [3] . The L278M mutant exhibited a 13-fold increase in half-life at 48˚C and a 12-fold higher T1/2 than WT-CALB [4] . Additionally, L278V mutant could improve enzyme enantioselectivity toward secondary alcohols [5] . All these results indicated that 278 site was important for the activity, enantioselectivity and thermo-stability of the enzyme. Beside these characterizations, substrate selectivity and catalytic mechanism are notable features of lipases and receive much concern by the enzyme engineers. However, distinct role of residue at 278 site played on the substrate specificity of the enzyme still could not acquire so for. It had been found that the shift of substrate specificity may be achieved by mutations on the lid or changing size and geometry of the substrate-binding cleft [6] [7] . Due to the low hit rate to get desired mutants by directed evolution, therefore, rational design is often used to modify proteins whose crystal structures or homolog structures are available.
Structural analysis on the crystal structures of the CALB (PDB ID: 1TCA) indicates that the side chain of leucine at 278 lies above the entrance of catalytic pocket, which prognosticates that this residue may play a role in substrate specificity of the enzyme. Herein, to further confirm this consumption, shortened side chain mutation of L278G and L278P was rational designed. The WT-CALB and mutant enzymes were purified to homogeneity in vitro and substrate specificity properties of these mutants were investigated. Subsequently, to further insight into the selectivity mechanism of WT-CALB and its mutants, models of mutants were constructed based on the crystal structure of CALB. Meanwhile, docking of various chain length substrates to the catalytic pocket of enzyme was proceeded to try to clarify the mechanism on substrate specificity. Beyond generating customized catalysts, these studies will help to elucidate the structure-function relationship of CALB.
2. Materials and Methods
2.1. Bacterial Strains, Chemicals, Plasmids and Medium
Plasmid pGAPZαA (Invitrogen) was used as expressing vector. Pichia pastoris X-33 (Invitrogen) strain was used for expression. The p-nitrophenol and p-nitrophenol esters with different chain length: p-nitrophenyl caprylate (pNP-C8), p-nitrophenyl caprate (pNP-C10), p-nitrophenyl laurate (pNP-C12) were purchased from Sigma Aldrich. pGAPZαA-CALB plasmid was constructed previously in our lab. Site-directed mutagenesis of CALB was carried out by the overlap extension method and using the pGAPZαA-CALB plasmid as template. The PCR products were double digested by KpnI/NotI and ligated into a linearized pGAPZαA vector. The mutations were confirmed by DNA sequencing and then linearized by restriction enzyme BlnI and transformed into P. pastoris X-33 strain by electroporation. The transformants were selected on YPD plates supplemented with Zeocin™ (25 μg/ml) and 3% (v/v) emulsified tributyrin at 30˚C.
2.2. Expression and Purification of Enzyme
The P. pastoris X-33 strains containing the recombinant plasmids were grown and expressed in YPD liquid medium (1% (w/v) yeast extract, 2% (w/v) peptone and 2% (w/v) glucose) at 30˚C with shaking of 200 rpm for 72 h. The supernatant of fermentation broth after centrifugation (10,000 g, 20 min, 4˚C) was filtered through a 0.22 μm filter membrane by suction filtration instrument (Autoscience AP-01P, China), then concentrated and buffer-exchanged to buffer A (20 mM phosphate buffer, pH 6.0, at 4˚C) through a 10 kDa molecular mass membrane (Vivaflow 200, Sartorius, Germany). The wild-type and mutants CALB were purified using DEAE ion- exchange chromatography. Buffer A that contain of crude enzyme was loaded into a chromatographic column and washed with buffer B (2.0 M NaCl in buffer A). The sample which flow through the column was collected, and then buffer-exchanged to phosphate buffered saline (PBS) (pH 7.4) and concentrated with a Macrosep® Advance Centrifugal Devices (10 kDa molecular weight cut-off) (Pall Life Science, USA). The purified lipase was analyzed by 15% SDS-PAGE. Protein concentrations were determined by the BCA Protein Assay Kit (Sangon Biotech Shanghai Co., Ltd).
2.3. Determination of Kinetic Constants
The kinetic parameters were determined by measuring the initial velocity of hydrolysis at different concentrations (0.01 - 1.0 mM) of p-nitrophenyl esters list above. The reaction buffer was PBS (pH 7.4) and reacted at 50˚C. The absorbance of the reaction mixture was measured at 405 nm. One unit of enzyme activity is defined as the amount of enzyme required to release 1 μmol of p-nitrophenol per minute. All the results in this study were the mean of triplicate measurements. The linear regression was applied to Lineweaver-Burk plot for estimation of Vmax, Km and kcat from the experimentally measured values. The Lineweaver-Burk plot has been widely used to determine important terms in enzyme kinetics, such as Km and Vmax. The y-intercept of such a graph was equivalent to the inverse of Vmax and the x-intercept of the graph represented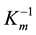 . Catalytic efficiency was determined by kcat/Km.
. Catalytic efficiency was determined by kcat/Km.
2.4. Construction of Mutant Models
To provide insight into the substrate selectivity mechanism of lipase CALB and its mutants, Discovery Studio V3.1 (Accelrys, San Diego, CA) was used for model construction. Models of mutants were constructed based on the crystal structure of lipase CALB (PDB ID: 1TCA) using the protocol Macromolecules-Design Protein- Modify structure-Build Mutants. The built models were solvated in explicit solvent and further minimized using the molecular dynamics simulation protocols to ensure the stability of the built models. The MD simulations were performed according to Liu et al. [8] . Docking experiments were performed by Glide 6.1 with default docking parameter settings according glide quick start guide.
3. Results and Discussion
3.1. Substrate Preference of WT-CALB and Its Mutants
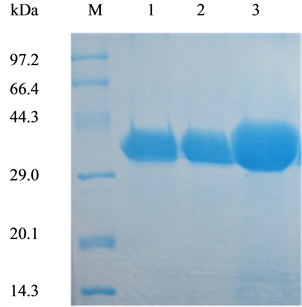
In this work, we succeeded in the heterologous production of extracellular WT-CALB and its two mutants. The WT-CALB and mutant lipases were purified through anion exchange chromatography. Subsequently, the purity of lipases was evaluated by 15% SDS-PAGE. The purified enzymes showed only a single band located at about 34 kDa according to the standard protein markers (Figure 1), coincidence with an apparent molecular weight reported before [9] [10] and can be used for further enzymatic studies.
Figure 1. SDS-PAGE analysis of wild-type CALB and two mutants produced in Pichia pastoris X-33. Lane M, molecular weight marker; lane 1, WT-CALB; lane 2, purified L278G mutant protein; lane3, purified L278P mutant protein. Ten microliter of appropriately diluted enzyme solutions was analyzed on 15% SDS-PAGE gels.
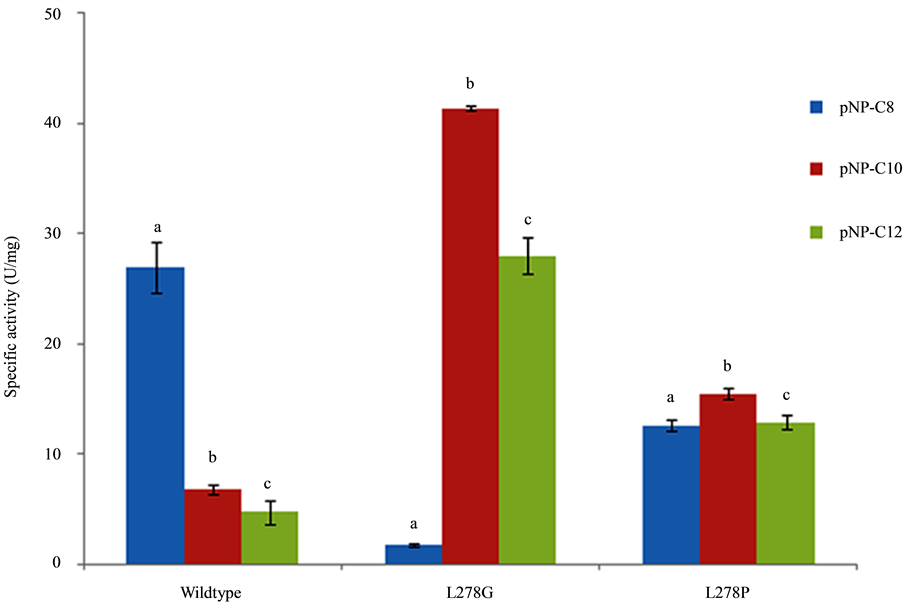
The colorimetric assay using p-nitrophenyl esters of fatty acids with various chain-lengths is often used to study the substrate preference of lipases. Specific activity of WT-CALB and mutants lipase toward various p-NP esters (C8-C12) were assayed using purified lipase. As shown in Figure 2, great difference in specific activity varying with the chain length of p-NP esters (C8-C12) was found for WT-CALB. WT-CALB shows a clear preference for pNP-C8 and low activity was found for pNP-C10 and pNP-C12. However, substrate specificity profiles of L278G and L278P mutants changed significantly. It is apparent that L278G and L278P all showed higher specific activity over the WT-CALB when with pNP-C10 or pNP-C12 as substrate. Among them, the L278G mutant had the highest specific activity to pNP-C10 and demonstrating nearly a 2-fold increase than WT-CALB (Figure 2). Conversely, the specific activity of the two mutants to pNP-C8 tested here all showed lower than WT-CALB. The effect of amino acid substitution on enzymatic activity exactly matches previously reported results [2] [11] .
3.2. Kinetic Constants of Purified CALB and Its Mutants
Initial velocity studies were performed to determine the kinetic parameters of the selected mutants using pNP- C8, pNP-C10 and pNP-C12 as substrate, respectively (Table 1). For the WT-CALB, the biggest affinity was found for pNP-C8, with Km value only was 69.30 μmol/L. Conversely, the Km value for pNP-C10, pNP-C12 was 5.7-fold and 3.7-fold higher than for pNP-C8. The kcat value decreased with the chain length increase, and the biggest value was found for pNP-C8. Similarly, the biggest catalytic efficiency (kcat/Km) value was found for pNP-C8. There was a great decrease in catalytic efficiency when with pNP-C10 and pNP-C12 as substrate.
For the L278G mutants tested here, the kcat value for pNP-C10 and pNP-C12 was higher than that of WT- CALB. The biggest kcat and Vmax value was both found for the pNP-C10, and was 2.2-fold and 1.5-fold higher than that of WT-CALB. Moreover, the catalytic efficiency of L278G for pNP-C10 and pNP-C12 was also found higher than that of WT-CALB. The biggest catalytic efficiency was found for pNP-C10 and was 1.9-fold higher than of WT-CALB. There was a great decrease in the catalytic efficiency to pNP-C8, with only 0.81 min−1・μM−1 remained, and significantly lower than the 14.28 min−1・μM−1 of WT-CALB.
Figure 2. Substrate specificity of L278G and L278P mutants toward p-nitrophenol esters of different acyl-chain length compared to that of wild-type CALB. The different letter represents significant difference between each groups (P < 0.05).
Compared with WT-CALB, L278P mutant did not change the affinity for pNP-C8 significantly. However, the kcat value of L278P for pNP-C10 and pNP-C12 was both higher than that of WT-CALB. For the L278P, the biggest kcat and Vmax value was found for pNP-C10, and was 2.4-fold and 7.4-fold higher than that of WT- CALB.
3.3. Modeling of L278G and L278P Mutants
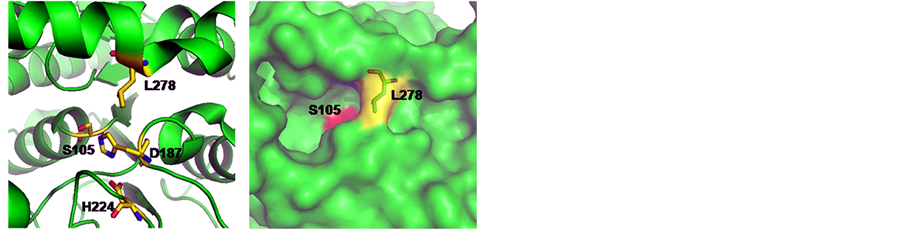
Structurally, CALB belongs to the α/β-hydrolase family and follows the same reaction mechanism as other serine hydrolases [12] . As can be seen from the 3D structure of WT-CALB (PDB:1TCA), the catalytic triad of CALB is made up of Ser 105, Asp187, and His 224, and the active-site channel is formed by three structural parts including helices α5, α10 and a loop region (Figure 3(a)). The C-terminal helix α10 (amino acids 268 to 287) is near the active site of the enzyme. It is dominated by alanine residues and is kinked in the middle at a proline residue (Pro280). L278 located at the α helix 10 which composed of the catalytic pocket, with its side chain direct to the center (Figure 3(a)), just like a wall blocked in the hydrophobic channel (Figure 3(b)). Moreover, in the crystal structure of WT-CALB, Leu278 exhibits a large chi angle deviation from ideality, indicating a distorted configuration [3] . Based on the X-ray crystal structure of the WT-CALB, docking calculations was performed for pNP esters tested here, and results revealed that while the smaller substrate, pNP-C8, fits tightly into the active site of the WT-CALB, the significantly larger substrate, pNP-C10 and pNP-C12, did not bind nearly as well due to steric hindrance interacting with the leucine side chain of the WT-CALB (Figure 4(a)). Therefore, even though the benzene ring of pNP-C10 could get into the catalytic pocket, it still hard to contact with the serine of catalytic traid. This structural observation was consistent with the observed increase in Km value for pNP-C10 and pNP-C12 of the WT-CALB.
To investigate the effects of L278G and L278P mutations on the CALB structure, homology modeling was used to construct the models of the mutants. After L278 was instead by glycine or proline, the side chain of the residues was greatly shortened and no space block existing above the catalytic pocket (Figure 3(c), Figure 3(d)).
Table 1. Kinetic parameters of purified wild-type CALB and its mutants for hydrolyzing of p-nitrophenol esters with different chain lengths.

Figure 3. 3D models of CALB mutants at residue 278 that generated by using the crystal structure of CALB (PDB: 1TCA) as the template. (a) The space position of catalytic triad and leucine 278 in wild-type CALB; (b) 3D Structure of wild-type CALB; (c) L278G mutant; (d) L278P mutant. Serine 105 was colored in red. Discovery Studio V3.1 (Accelrys, San Diego, CA) was used for model construction. Models of mutants were constructed based on the crystal structure of lipase CALB (PDB ID: 1TCA) using the protocol macromolecules-design protein-modify structure-build mutants. The built models were solvated in explicit solvent and further minimized using the molecular dynamics simulation protocols to ensure the stability of the built models.
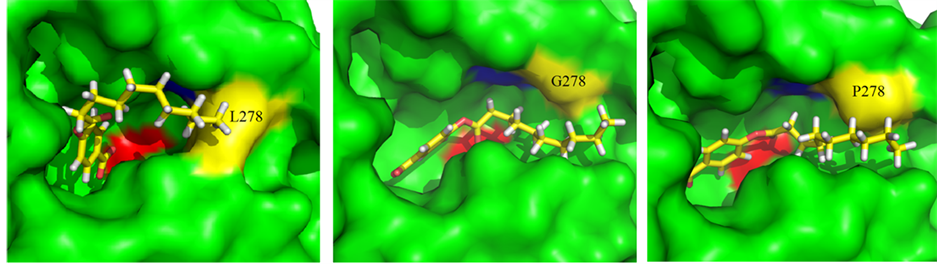
Figure 4. Substrate models within the active site (Serine 105 is colored in red) of WT-CALB, L278G and L278P. (a) pNP-C10/WT-CALB; (b) pNP-C10/L278G; (c) pNP-C10/L278P.
Meanwhile, no significant differences in the models were found before and after the MD simulations. A decrease in van der Waals volume of the mutated residue provide a subtle change in the active site pocket, which have a larger space for the substrate [3] . The larger channel to the active site could allow bigger substrate enter to the catalytic pocket and closer conjunct with pNP-C10 than pNP-C8 was found as illustrated by docking with pNP-C10 (Figure 4(b), Figure 4(c)). This local change in the active site, may contribute to the increase in the lipase activity, corresponding to the increase of kcat without significant effect on Km, and finally induced the catalytic efficiency of the two mutants for pNP-C10 and pNP-C12 substrates exceeded that of the wild-type enzyme.
4. Conclusion
In conclusion, improvement substrate specificity of CALB to longer chain acyl ester by amino acid substitution of leucine 278 with glycine or proline was found in present study. It was mainly the stretch of side chain leucine 278 above the catalytic pocket that resulted in the shorter chain length selectivity. The shortened side chains of glycine or proline break the steric hindrance existing above the catalytic pocket and fit well with the longer chain esters, and thus make it display longer chain preference. However, only two kinds of amino acid substitution were constructed and investigated in the present study. Further research still needs to do to clarify the influence of other amino acid substitution on this site to the substrate specificity of the enzyme.
Acknowledgements
This work was made possible with funding provided by the National Natural Science Foundation of China (No.31401627, No.21376098), and Science and Technology Planning Project of Guangdong Province, China (No. 2013B051000009), and the Fundamental Research Funds for the Central Universities (No. 2014ZM0062).
References
- Juhl, P.B., Doderer, K., Hollmann, F., Thum, O. and Pleiss, J. (2010) Engineering of Candida antarctica Lipase B for Hydrolysis of Bulky Carboxylic Acid Esters. Journal of Biotechnology, 150, 474-480. http://dx.doi.org/10.1016/j.jbiotec.2010.09.951
- Park, C.G., Kwon, M.A., Song, J.K. and Kim, D.M. (2011) Cell-Free Synthesis and Multifold Screening of Candida antarctica Lipase B (CalB) Variants after Combinatorial Mutagenesis of Hot Spots. Biotechnology Progress, 27, 47- 53. http://dx.doi.org/10.1002/btpr.532
- Kim, S.Y., Sohn, J.H., Pyun, Y.R., Yang, I.S., Kim, K.H. and Choi, E.S. (2007) In Vitro Evolution of Lipase B from Candida antarctica Using Surface Display in Hansenula polymorpha. Journal of Microbiology and Biotechnology, 17, 1308-1315.
- Xie, Y., An, J., Yang, G., Wu, G., Zhang, Y., Cui, L. and Feng, Y. (2014) Enhanced Enzyme Kinetic Stability by Increasing Rigidity within the Active Site. The Journal of Biological Chemistry, 289, 7994-8006. http://dx.doi.org/10.1074/jbc.M113.536045
- Marton, Z., Léonard-Nevers, V., Syrén, P.-O., Bauer, C., Lamare, S., Hult, K., Tranc, V. and Graber, M. (2010) Mutations in the Stereospecificity Pocket and at the Entrance of the Active Site of Candida antarctica Lipase B Enhancing Enzyme Enantioselectivity. Journal of Molecular Catalysis. B, Enzymatic, 65, 11-17. http://dx.doi.org/10.1016/j.molcatb.2010.01.007
- Santarossa, G., Lafranconi, P.G., Alquati, C., DeGioia, L., Alberghina, L., Fantucci, P. and Lotti, M. (2005) Mutations in the “Lid” Region Affect Chain Length Specificity and Thermostability of a Pseudomonas fragi Lipase. FEBS Letters, 579, 2383-2386. http://dx.doi.org/10.1016/j.febslet.2005.03.037
- Yang, J., Koga, Y., Nakano, H. and Yamane, T. (2002) Modifying the Chain-Length Selectivity of the Lipase from Burkholoderia cepacia KWI-56 through in Vitro Combinatorial Mutagenesis in the Substrate-Binding Site. Protein Engineering, 15, 147-152. http://dx.doi.org/10.1093/protein/15.2.147
- Liu, L., Gao, C.L., Lan, D.M., Yang, B. and Wang, Y.H. (2012) Molecular Basis for Substrate Selectivity of a Mono- and Diacylglycerol Lipase from Malassezia globosa. Biochemical and Biophysical Research Communications, 424, 285-289. http://dx.doi.org/10.1016/j.bbrc.2012.06.108
- Larsen, M.W., Bornscheuer, U.T. and Hult, K. (2008) Expression of Candida antarctica Lipase B in Pichia pastoris and Various Escherichia coli Systems. Protein Expression and Purification, 62, 90-97. http://dx.doi.org/10.1016/j.pep.2008.07.012
- Eom, G.T., Lee, S.H., Song, B.K., Chung, K.W., Kim, Y.W. and Song, J.K. (2013) High-Level Extracellular Production and Characterization of Candida antarctica Lipase B in Pichia pastoris. Journal of Bioscience and Bioengineering, 116, 165-170. http://dx.doi.org/10.1016/j.jbiosc.2013.02.016
- Suen, W.C., Zhang, N., Xiao, L., Madison, V. and Zaks, A. (2004) Improved Activity and Thermostability of Candida antarctica Lipase B by DNA Family Shuffling. Protein Engineering, Design & Selection, 17, 133-140. http://dx.doi.org/10.1093/protein/gzh017
- Wu, Q., Soni, P. and Reetz, M.T. (2013) Laboratory Evolution of Enantiocomplementary Candida antarctica Lipase B Mutants with Broad Substrate Scope. Journal of the American Chemical Society, 135, 1872-1881. http://dx.doi.org/10.1021/ja310455t
NOTES

*Corresponding author.