Open Journal of Medical Microbiology
Vol.2 No.2(2012), Article ID:19633,4 pages DOI:10.4236/ojmm.2012.22005
Characterization of Aeromonas hydrophila through RAPD-PCR and SDS-PAGE Analysis
1Department of Biotechnology, The University of Burdwan, Burdwan, India
2Department of Biotechnology, Oriental Institute of Science and Technology, Burdwan, India
3Central Inland Fisheries Research Institute, Barrackpore, India
Email: *agniswar.sarkar@yahoo.co.in
Received December 29, 2011; revised January 30, 2012; accepted February 20, 2012
Keywords: RAPD-PCR; SDS-PAGE; Aeromonas hydrophila; Molecular Characterization
ABSTRACT
Aeromonas hydrophila isolated from fish (Labeo rohita), pond water, river water, raw meat of chicken and mutton and raw cow milk were characterized through Random Amplified Polymorphic DNA (RAPD) analysis and Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS PAGE) of cellular proteins. RAPD-PCR amplification of bacterial DNA was done by using ten random primers (OPA-01 to 10) and found some distinct banding pattern on agarose gel. RAPD profile was studied with each isolate and absolute polymorphism indicating its application as an ideal tool for molecular characterization. Other methods like morphological, serological and biochemical characterization gives contradictory results and total cellular protein profiling does not show any significant polymorphism for identification and discrimination.
1. Introduction
Aeromonas hydrophila are Gram-negative, non-sporeforming and rod-shaped to coccoid cell with rounded ends. They are oxidase and catalase positive, produce nitrate to nitrite and ferment D-glucose. A. hydrophila have their natural habitat in water and grow over a wide temperature range between 0˚C and 45˚C, with a temperature optimum of 22˚C to 32˚C [1-3]. They have been isolated from water especially in surface water and sewage. They also occur in untreated and treated drinking water, soil and foodstuffs [4]. This species are responsible for severe haemorrhagic syndrome in a variety of fishes and multiple diseases in poikilothermic animals [5]. This species are shown to be potentially pathogenic and associated with several human infections, including gastrointestinal infections and extra-intestinal infections, such as endocarditis, meningitis, septicaemia and urinary tract and wound infections. Abscesses or wound infections associated with exposure to soil or water represent the most prevalent extra-intestinal infections [6,7]. Aeromonas spp. are found to be serologically heterogeneous, with individual serogroups found in more than one species. Most type and reference strains were not serologically representative of a genomospecies. Various genotypic typing methods have been applied for identification to this species [7-9]. Plasmid analysis is unhelpful because plasmid carriage is infrequent (20% - 58%) in A. hydrophila [7]. In contrast, rRNA gene restriction patterns provide good discrimination within A. hydrophila [8,9]. In the present paper, the importance Aeromonas identification and use of molecular genotyping methods as RAPD analysis has been elaborated and cellular proteins also compared with the RAPD data and given some unique pattern.
2. Materials and Methods
2.1. Isolation of Bacteria and Genomic DNA Extraction
Thirty isolates of Aeromonas hydrophila from different sources like fish (Labeo rohita), raw cow milk, pond water, river water, raw meat of chicken and mutton in West Bengal, India were used in this study. Aeromonas hydrophila were isolated by using a selective medium, Rimler Shott agar (Hi Media). The plates were incubated at 37˚C for 28 hours. All cultures were identified to the species level using Automated Microbial Analyzer (Biolog, US). Selected Aeromonas hydrophila colonies were subcultured in Tryptic Soya Broth (Difco) for further characterization.
By following the protocol of Sambrook, 1889 [10], DNA was isolated by modification of overnight grown bacterial culture in 5 ml TSB and centrifuge at 12,000 g × 5 min at 4˚C. Resuspended pellet in 0.2 ml of TE-1 buffer and added 50 µl lysozyme (3 mg/ml) and keep at 37˚C × 15 min then 50 µl lysozyme was added with 50 µl proteinase K (15 mg/ml) and incubated at 56˚C × (3 - 4) hrs in a water bath. DNA were extracted by adding equal volume of TE saturated phenol by slow mixing and centrifugation 12,000 g × 5 min at 4˚C. With the supernatant, equal volume of phenol: chloroform: iso-amyl alcohol (25:24:1) was added and again centrifuged. To the supernatant, 200 µl chloroform was added and supernatant was collected after centrifugation and 25 µl of 5 M NaCl. 250 µl of chilled absolute alcohol was added and DNA was observed as bunch of threads. Then DNA was collected after wash with 70% alcohol and dried it. Pure DNA was dissolved in TE buffer and stored at 4˚C for further use.
Total ten numbers of random primers, designated as OPA-1 to OPA-10 was used for PCR amplification. Out of these 10 primers, viz., OPA-03, OPA-09 and OPA-10 were selected for final screening because they only generated several reproducible amplicons and produced some distinct banding patterns on 1.5% agarose gel slab. In the present paper result of OPA-03 has been elaborated.
2.2. PCR Amplification and Analysis of Phylogeny
PCR reactions was standardized by a series experiments and finally carried out by following the protocol of P. C. Thomas, 2009 [11], for the annealing temperature, concentration of MgCl2, template DNA, Taq DNA polymerase, dNTP’s and primers. The PCR reaction components consists of 200 µM dNTP, 20 pmoles of each primer, 2 units of Taq DNA polymerase, assay buffer with 1.5 mM MgCl2, 20 - 30 ng template DNA in an assay volume of 25 µL.
PCR reaction was performed with Thermo Electron PCR system. Initial denaturation at 94˚C × 5 min followed by 35 cycles at 94˚C × 1 min, 36˚C × 1 min and 72˚C × 2 min. The final extension was done at 72˚C × 10 min. PCR amplicons were analyzed on 1.5% agarose gel.
Amplified DNA bands on the 1.5% agarose gel were analyzed by giving scores of zero and one for the absence or presence of bands at each band position. The similarity index between isolates was calculated by Nei and Li’s protocol, 1979 [12]. Genetic similarity between isolates: A & B (SAB) was calculated using the formula: –SAB = 2NAB/(NA + NB). Where SAB = Genetic similarity between A & B. NAB—Number of amplified bands shared in common between isolate A & B. NA and NB—Total number of bands possessed by the isolates A & B, respectively. Further cluster analysis was performed using this matrix in SAS programme to create a phenogram. Statistical analysis was carried out by one way analysis of variance (ANOVA) in SAS (version 6.12) to test the level of significance.
2.3. Extraction of Bacterial Protein and SDS-PAGE
Aeromonas hydrophila were inoculated in 5 ml Tryptic soya broth for 28 hours at 37˚C and cells were collected and centrifuged at 10,000 g × 10 min at 4˚C. The supernatant was drained off and 100 µl of B-PER reagent (Bacterial Protein Extraction Reagent) was added to each of the pellets and vortexed. This was incubated at 4˚C for 15 minutes, mixing gently every five minutes. The cell suspension was then centrifuged at 10,000 g × 10 min at 4˚C and the supernatant was collected in eppendorf tubes which contains exocellular proteins and pellets were dissolved with 100 µl of PBS and vortexed, which contain cellular proteins and stored at –20˚C for further use. 12% SDS-PAGE was carried out in the present study for the total proteomics analysis.
3. Results
Aeromonas hydrophila produced yellow colonies in the RS-medium. Gram staining of these colonies gives gram negative reaction, microscopically analysis gives rod shaped, motile, biochemical tests gives oxidase positive, fermentative and antibiotic resistance tests concluded as novobiocin resisitant, primarily indicated that colonies are aeromonads. All isolates were confirmed to the species level Aeromonas hydrophila by Automated Microbial Analyzer (Biolog, US).
3.1. Analysis of RAPD Profile
The RAPD-PCR carried out by following of each cycle of initial denaturation at 94˚C × 5 min followed by 35 cycles at 94˚C × 1 min, 36˚C × 1 min and 72˚C × 2 min. The final extension at 72˚C × 10 min PCR amplified products were analyzed on 1.5% agarose gel slab and it produced clear reproducible bands. Amplification of the DNA from each isolates produced a variety of amplicons with distinct bands on 1.5% agarose gel after electrophoresis. The RAPD fingerprints of the isolates generated by these random primers OPA-03 is given in Figure 1. In this study, The RAPD fingerprint pattern was more or less unique for each isolates and each banding pattern indicates that there were a number of fragments, which were also homogenous between many of the isolates. These unique fragments may be utilized for the development of species-specific marker.
Scanning of RAPD gel gives total of 70 bands where some scorable as distinct bands in the gel. Whereas many of the amplicons were produced by all the isolates, some of amplicons showed variation between isolates. Polymorphism of the RAPD pattern was quite apparent among the different isolates. The RAPD fingerprint pattern was unique for each of the isolates. Comparison of the amplicons at each loci indicated there were variety of polymorphism. However, banding pattern indicates that there were a number of fragments, which were homogenous among many of the isolates. The analysis indicated that the overall polymorphism of these fragments may be utilized for species-specific marker development and other aeromonads can also be examined with these primers.
3.2. Analysis of Cellular Protein
Cellular proteins were extracted from the isolates and analyzed through 12% SDS-PAGE technique (Figure 2). No significant polymorphism was found (Other than in lane 4). Banding patterns of all the isolates were more or less uniform. Molecular weight marker was used to identify the molecular weight and density of each band of each lane in the gel of all the isolates in the study.
4. Discussion
A. hydrophila are phenotypically, serologically and genetically quite diverse, many conventional methods of identifying these microorganisms like cultural-biochemical properties and serotyping give contradictory results compared to the molecular tools. Because of the complexity of methodologies, time taking and difficult interpretation of these results, genomic analysis methods have been commonly employed to characterize the microbial pathogens. All the isolates were typable using selected primers. But while performing screening for 10 primers, only primer 09 amplified with scorable bands and others had very poor reproducibility.
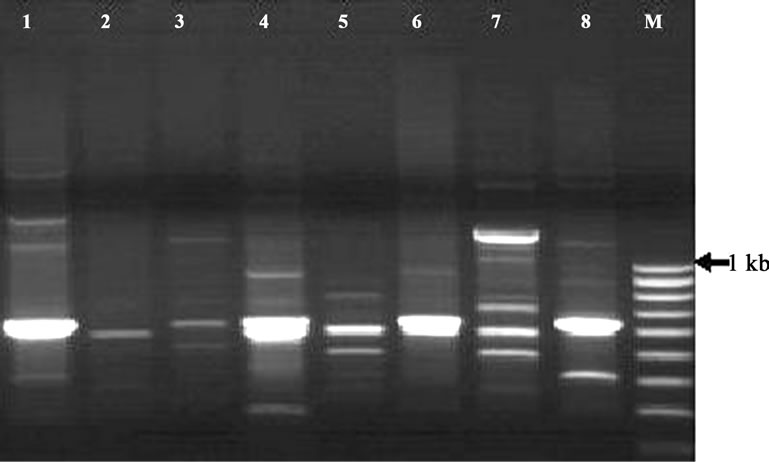
Figure 1. RAPD profile of Aeromonas hydrophila isolates generated by primer OPA-03 using random primer and showing variable polymorphisms and unique DNA bands. Lane M: 100 bp ladder (Image analyzed by Gel Doc System, 2000, Biorad).
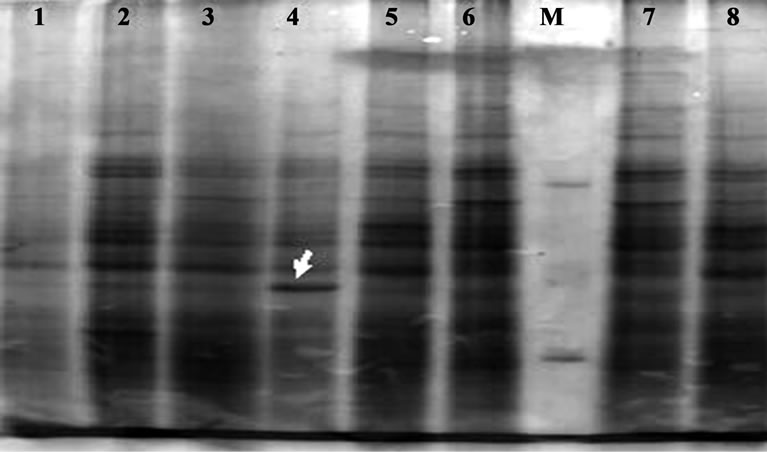
Figure 2. Cellular protein profile of A. hydrophila resolved through SDS-PAGE (12% resolving gel and 5% stacking gel) and stained with coomassie brilliant blue r250. Lane 1-08: Ah1-Ah08 isolates; M: M. W. Marker.
The technique being simple, specific and cost effective is being widely used as an alternate to other fingerprinting methods. Requirement of very small amount of DNA, without any prier information on genomic DNA sequence and use of universal primers make RAPD-PCR a popular DNA fingerprinting method in genomic analysis. Miyata, 1995 [13] observed that the DNA required for RAPD is less than one hundredth of the amount required for other methods. This study also performed to generate reproducible RAPD profiles with as little amount of DNA in the PCR reaction in the molecular characterization of A. hydrophila strains from different sources of West Bengal, India. Results of this study established the observation by other workers that motile aeromonads are genetically diverse [14]. RAPD-PCR fingerprints have been used for typing and diagnosis of bacteria. So, this molecular typing method could be used as a new strategy for epidemiological investigations. This information can be used to improve quality control and bio-security protocols to check Aeromonas disease outbreaks and this concept can be applied to other bacterial pathogens.
5. Conclusion
RAPD marker could be used as molecular markers of A. hydrophila pointed towards the amplicons generated by OPA-03. The species specificity can be confirmed only by checking these primes with isolates of other aeromonads. Large number of RAPD genotypes can be an ideal method for species identification [15]. The results show that RAPD fingerprinting is one of the best molecular tools for identification but protein profiling has some limits to differentiate Aeromonas species, and the present study indicates that SDS-PAGE technique is not suitable for the characterization within a species.
6. Acknowledgements
The authors thank to Head of Department, Department of Biotechnology, The University of Burdwan, Burdwan, West Bengal, 713104 for providing necessary facilities and intellectual input. The author also acknowledges UGC for the financial assistance received during the tenure of this work. Dr. P. Roy wishes to acknowledge Shri Mrigendra Nath Mondal endowment Chair for Professor in Biotechnology. Authors also acknowledge to Avijit Patra for his contribution in Computational Biological work.
REFERENCES
- M. Altwegg, “Subtyping Methods for Aeromonas Species,” In: B. Austin, M. Altwegg, P. J. Gosling and S. W. Joseph, Eds., The Genus Aeromonas, John Wiley & Sons, Chichester, 1996, pp. 109-126.
- M. Altwegg, “Aeromonas and Plesiomonas,” In: P. Murray, E. Baron, M. Pfaller, F. Tenover and R. Yolken, Eds., Manual of Clinical Microbiology, ASM Press, Washington DC, 1999, pp. 507-516.
- G. Brandi, M. Sisti, G. F. Schiavano, L. Salvaggio and A. Albano, “Survival of Aeromonas hydrophila, Aeromonas caviae and Aeromonas sobria in Soil,” Journal of Applied Microbiology, Vol. 81, No. 4, 1996, pp. 439-444. doi:10.1111/j.1365-2672.1996.tb03531.x
- M. Kupfer, P. Kuhnert, B. M. Korczak, R. Peduzzi and A. Demarta, “Genetic Relationships of Aeromonas Strains Inferred from 16S rRNA, gyrB and rpoB Gene Sequences,” International Journal of Systematic and Evolutionary Microbiology, Vol. 56, No. 12, 2006, pp. 2743-2751. doi:10.1099/ijs.0.63650-0
- M. Altwegg, H. K. Geiss and B. J. Freij, “Aeromonas as a Human Pathogen,” Critical Reviews in Microbiology, Vol. 16, No. 4, 1989, pp. 253-286. doi:10.3109/10408418909105478
- R. D. Isaacs, S. D. Paviour, D. E. Bunker and S. D. R. Lang, “Wound Infection with Aerogenic Aeromonas Strains: A Review of Twenty-Seven Cases,” European Journal of Clinical Microbiology & Infections Diseases, Vol. 7, No. 3, 1988, pp. 355-360. doi:10.1007/BF01962336
- J. M. Janda, “Recent Advances in the Study of the Taxonomy, Pathogenicity, and Infectious Syndromes Associated with the Genus Aeromonas,” Clinical Microbiology Reviews, Vol. 4, No. 4, 1991, pp. 397-410.
- E. J. Kuijper, L. van Alphen, E. Leenders and H. C. Zanen, “Typing of Aeromonas Strains by DNA Restriction Endonuclease Analysis and Polyacrylamide Gel Electrophoresis of Cell Envelopes,” Journal of Clinical Microbiology, Vol. 27, No. 6, 1989, pp. 1280-1285.
- N. P. Moyer, G. Martinetti, J. Luthy-Hottenstein and M. Altwegg, “Value of rRNA Gene Restriction Patterns of Aeromonas spp. for Epidemiological Investigations,” Current Microbiology, Vol. 24, No. 1, 1992, pp. 15-21. doi:10.1007/BF01570094
- J. Sambrook, E. F. Fritsch and T. Maniatis, “Molecular Cloning: A Laboratory Manual,” 2nd Edition, Cold Spring Harbor Laboratory, New York, 1989.
- P. C. Thomas, P. R. Divya, V. Chandrika and M. P. Paulton, “Genetic Characterization of Aeromonas hydrophila Using Protein Profiling and RAPD PCR,” Asian Fisheries Science, Vol. 22, No. 2, 2009, pp. 763-771.
- M. Nei and W. H. Li, “Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucleases,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 76, No. 10, 1979, pp. 5269-5273.
- M. Miyata, T. Aoki, V. Inglis, T. Yoshida and M. Endo, “RAPD Analysis of Aeromonas salmonicida and Aeromonas hydrophila,” Journal of Applied Microbiology, Vol. 79, No. 2, 1995, pp. 181-185. doi:10.1111/j.1365-2672.1995.tb00933.x
- J. I. MacInnes, T. J. Trust and J. H. Corsa, “Deoxyribonucleic Acid Relationships among Members of the Genus Aeromonas,” Canadian Journal of Microbiology, Vol. 25, No. 5, 1979, pp. 579-586. doi:10.1139/m79-083
- H. Hadrys, M. Balick and B. Schierwater, “Applications of Random Amplified Polymorphic DNA (RAPD) in Molecular Ecology,” Molecular Ecology, Vol. 1, No. 1, 1992, pp. 55-63. doi:10.1111/j.1365-294X.1992.tb00155.x
NOTES
*Corresponding author.
#Authors’ contribution: The research work is really handled and designed by Agniswar Sarkar. Part of Computational work was handled by Agniswar Sarkar and Mousumi Saha. Mousumi Saha also helped him to conclude and finished the work and Pranab Roy also contributes his intellectual input for the study.

