Open Journal of Blood Diseases
Vol. 2 No. 2 (2012) , Article ID: 20186 , 5 pages DOI:10.4236/ojbd.2012.22004
JAK2 Mutations in Chronic Myeloproliferative Neoplasm; Towards the Application of Personalized Treatments for Saudi Patients

1Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, KSA; 2Center of Excellence in Genomic Medicine Research, King Abdulaziz University, Jeddah, KSA; 3Department of Hematology, Faculty of Medicine, King Abdulaziz University, Jeddah, KSA; 4Clinical Hematology Laboratory, King Abdulaziz Hospital and Oncology Center, Jeddah, KSA.
Email: *mgari@kau.edu.sa
Received January 24th, 2012; revised March 1st, 2012; accepted March 15th, 2012
Keywords: Leukemia; CMPN; PV; ET; PMF; JAK2 Mutation
ABSTRACT
The chronic myeloproliferative neoplasms (CMPN) are a group of clonal hematopoietic stem cell disorders in which large numbers of red blood cells, white blood cells, or platelets grow and spread excess in the bone marrow and the peripheral blood. Cytogenetic analysis of the t(9;22) and molecular detection of BCR/ABL is the main diagnostic criteria in Philadelphia positive CMPN (CML). The identification of non-receptor tyrosine kinase JAK2 mutations (exon 14 JAK2 V617F and exon 12) have significantly contributed to our understanding of the molecular mechanisms in the pathogenesis of Philadelphia negative CMPN such as polycythemia vera (PV), essential thrombocythemia (ET) and primary myelofibrosis (PMF) patients. According to the revised WHO classification, JAK2 mutation is considered as a major diagnostic and clonal marker in Philadelphia negative CMPN which will play a major role in designing personalized treatments for the disease. JAK2 V617F mutation frequency is unknown in Saudi Arabia. Therefore, investigation of the JAK2 V617F mutation was carried out in DNA samples from 78 peripheral blood specimens corresponding to patients with polycythemia vera (PV) (n = 11), Chronic myeloid leukemia (CML) (n = 45), essential thrombocythemia (ET) (n = 10), idiopathic myelofibrosis (MF) (n = 12). We used polymerase chain reaction and direct DNA sequencing to detect the JAK2 mutation. Overall, the incidence of the JAK2 V617F mutation was 91% in PV, 40% in ET, and 25% in MF. This approach proved to be reliable and more sensitive in detecting the mutation. Two essential findings arose from our study. First, this technique could be carried out with DNA samples, even partially degraded, from routinely processed BM or peripheral blood specimens. Second, after correlation with morphological features, it turned out that the characteristics of the megakaryocytes were more specific than the mutational status of JAK2 in characterizing ET and PMF. Concerning PV, as expected, the incidence of the JAK2 mutation was higher, but the morphological criteria were misleading in some cases, strongly suggesting that the combination of both morphology and molecular data would enable the characterization of virtually all cases. JAK2 V617F mutation frequency along with accurate morphological characterization is very reliable tool in diagnosing and classifying CMPN in Saudi patients.
1. Introduction
Chronic myeloproliferative neoplasms (CMPN) includeing chronic myelogenous leukemia (CML), polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF), are a phenotypically diverse category of malignancies that are thought to derive from stem cells in the myeloid lineage [1]. CMPN is characterized by an overproduction of mature myeloid cells, bone marrow hypercellularity, extramedullary hematopoiesisa tendency for thrombosis and or hemorrhage, and may sometimes progress to a leukemic transformation. Characterization of the molecular lesions underlying CMPN has broadened our understanding of the disease mechanism and has advanced diagnostic, prognostic, and therapeutic applications in this disease class. The first genetic alteration identified in CMPN was the translocation (9;22) that creates the BCR-ABL fusion gene resulting in a constitutively activated tyrosine kinase that is responsible for the activation of signal transduction pathways leading to CML [2,3]. The identification and characterization of the BCR-ABL chimeric protein led to the development of small molecules that target the tyrosine kinase activity of the chimeric protein. The genetic basis of Philadelphia negative CMPN was not clear until the year 2005 when it was first demonstrated that an acquired somatic mutation in the Janus kinase 2 gene (JAK2), causes a valine to phenylalanine substitution at position 617 (JAK2 V617F) resulting in a constitutively activated tyrosine kinase in most cases of CMPN [4-6]. In addition, the JAK2 gene is located on the short (p) arm of chromosome 9 at position 24. [7] More precisely, the JAK2 gene is located from base pair 4,985,244 to base pair 5,128,182 on chromosome 9 [7]. However, JAK2-STAT signaling pathway is essential for mediating cytokine dependent proliferation, survival and apoptosis, a disruption in this pathway in hematopoietic stem cells lead to myeloproliferative diseases. Activating mutations in JAK2 have been found in the majority of BCR-ABL-negative myeloproliferative neoplasms. The JAK2 (V617F) mutation has been previously reported in approximately 95% of patients with PV, 23% - 57% in ET and in 35% - 50% in PMF [8-10]. The identification of activating JAK2 mutations has raised considerable interest in the development of JAK2 inhibitors for the treatment of Philadelphia negative CMPN [11]. The aim of the current study was to determine the frequency of JAK2 mutations in the CMPN patients from Saudi Arabia.
2. Materials and Methods
2.1. Patients and Samples
The current study includes 78 patients diagnosed with CMPN collected at King Abdul Aziz University Hospital and King Abdulaziz Hospital and Oncology Center in the Western Region of Saudi Arabia between 2001 and 2010. The study population included 45 CML cases, 10 ET cases, 12 MF cases and 11 PV cases diagnosed using WHO 2001 classification [12]. Peripheral blood (PB) or bone marrow (BM) or bone marrow smear (BMS) were collected in accordance with institutional guidelines and conformed to the Helsinki Declaration [13]. Local ethical guidelines were adhered to throughout the sample collection process (ethical approval number 09-CEGMR-ETH).
2.2. DNA Preparation and Polymerase Chain Reaction
High-molecular-weight DNA was prepared from BM and PB using DNeasy Blood & Tissue Kit (Qiagen, 69504) according to manufacturer’s instructions. DNA preparation from BMS was prepared using the same protocol however the preparation of the starting material was done as follows: the bone marrow slides were scraped and the smear was collected in a 1.5 ml tube, followed by dissolving in 100 µl phosphate buffered saline and heating at 55˚C for 15 min, followed by DNA preparation using DNeasy Blood & Tissue Kit. The entire coding sequence and intron/exon boundaries corresponding to exons 12, 13 and 14 of the JAK2 gene were amplified independently, using the following primers: Exon 12 forward (5’-CAAAGTTCAATGAGTTGACCCCTA-3’) and reverse (5’-CCAATGTCACATGAATGTAAATCAA-3’); Exon 13 forward (5’-TCTTGTTCCTACTTCGTTCTCCA-3’) and reverse (5’-TTGAAAAGCTGCACACATGA-3’) and Exon 14 forward (5’-TCCTCATCTATAG TCATGCTG AAA-3’) and reverse (5’-CTGACACCTAGCTGTGA TCCTG-3’). The DNA-PCR assay was performed on a mixture of 25 µL containing 100 ng genomic DNA with 12.5 µL KAPA Taq ReadyMix (Kapa Biosystems) and 1µM each forward primer and reverse primer. The primer sequence for the 3 exons are are shown in Table 1. PCR was performed in an Matercycler® Pro(Eppendorf) with the following schedule: 3130An initial preheating at 94˚C for 5 minutes was followed by denaturation at 94˚C for 30 seconds, 60˚C for 30 seconds, and 72˚C for 30 seconds for a total of 35 cycles, and a final extension at 72˚C for 10 minutes. The PCR products were analyzed by electrophoresis on 2% agarose gels (Sigma), stained with ethidium bromide, and visualized under an ultraviolet lamp. The PCR product was purified using MinElute PCR Purification Kit (Qiagen), and directly sequenced in both directions with the BigDye Terminator Cycle Sequencing Ready Reaction kit, which contained AmpliTaq DNA polymerase FS (Applied Biosystems) on an automated ABI PRISM 3130 DNA sequencer (Applied Biosystems) according to the manufacturer’s instructions.
2.3. Determination of BCR-ABL Rearrangements
RT-PCR was used to detect common BCR/ABL fusion transcripts. Mononuclear cells were isolated from PB or BM of CMPN cases using Ficoll-Paque™ PLUS (GE Healthcare). RNA was prepared from the mononuclear cells using RNeasy Mini Kit (Qiagen). cDNA was prepared using QuantiTect Reverse Transcription Kit (Qiagen) and subjected to a multiplex RT-PCR as described before [14].
3. Results
Patient characteristics are shown in Table 1. The current study included 78 CMPN patients which comprises of 45 CML cases (57%), 10 ET cases (13%), 12 PMF cases (15%) and 11 PV cases (14%) (Figure 1). Median age at diagnosis was 60 years. Patients were diagnosed and categorized using WHO 2001 criteria [15]. The basic investigations included complete blood count, arterial
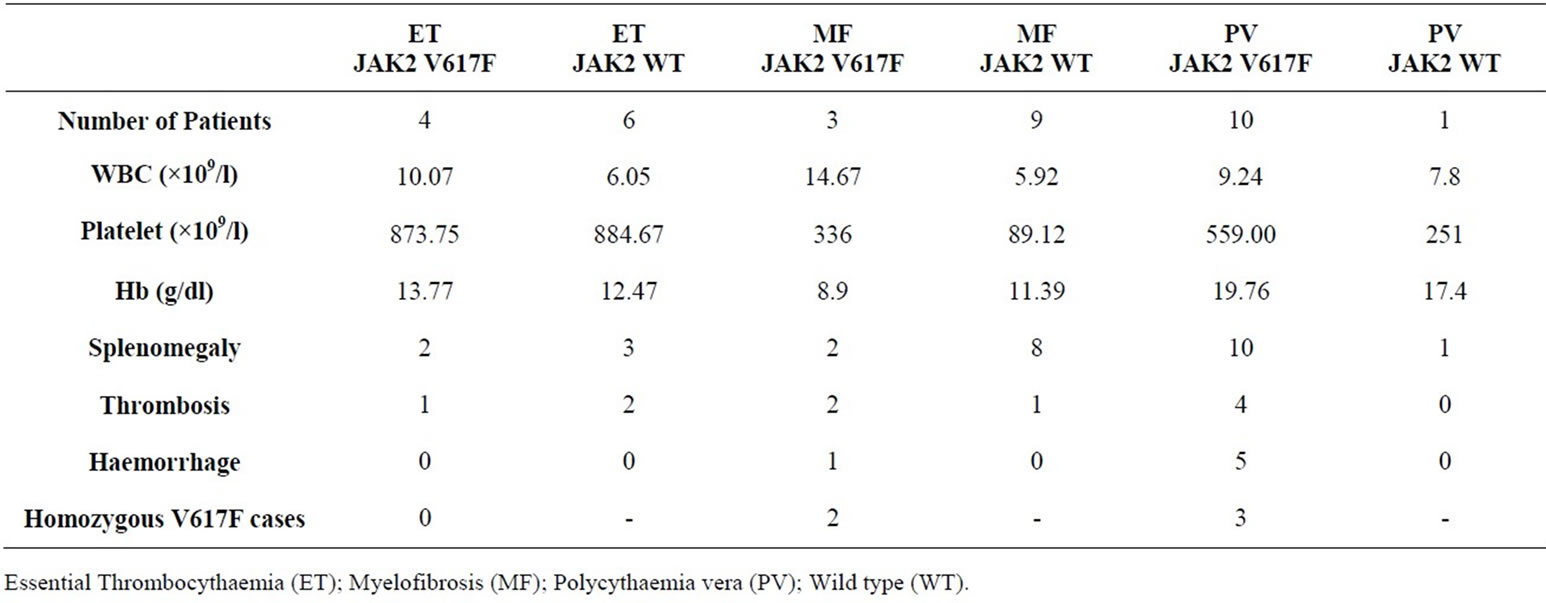
Table 1. Clinical characteristics of Ph-negative CMPN patients and their JAK2 status.
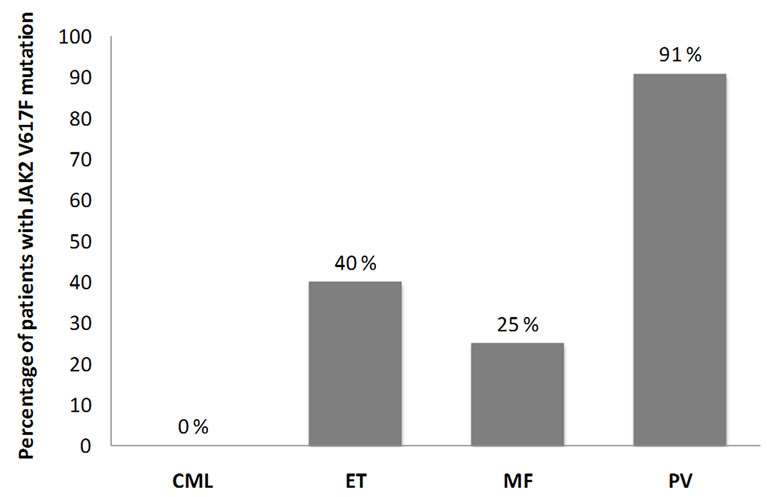
Figure 1. Percentage of patients with JAK2V617F in each subtype of CMPN; Chronic myelogenous leukemia (CML), Essential Thrombocythaemia (ET), Myelofibrosis (MF), Polycythaemia vera (PV).
oxygen saturation; serum erythropoietin levels and bone marrow aspiration and biopsy were routinely performed. The CML cases were confirmed based on karyotyping and/or molecular analysis of the suspected cases. 24 of the 45 CML cases were analyzed in our laboratory for the presence of translocation t(9;22) (Philadelphia chromosome) and/or molecular evidence of BCR/ABL fusion transcripts. Sequence analysis of the JAK2 exon 12, 13 and 14 were carried out on all 78 CMPN cases. Our results revealed JAK2 V617F mutation in 10 PV cases (91%), 4 ET cases (40%), 3 MF cases (25%) and none CML cases (0%). These results are in agreement with the previously reported JAK2 mutation frequencies in CMPN. A comparison of clinical characteristics between patients with JAK2 V617F mutation and those without is reported in Table 1.
Sequence analysis revealed 5 cases (29%) with homozygous JAK2 V617F mutation among all cases V617F positive cases. Figure 2 shows sequences of CMPN patients with wild type JAK2, heterozygous and homozygous V617F mutations.
4. Discussion and Conclusion
The association of JAK2 V617F (exon 14 mutation) and CMPN such as PV, ET and PMF was first reported in 2005 [4,16]. This discovery was soon followed by a number of studies reporting the incidence of JAK2 V617F mutation in CMPN [5-6,8]. The identification this activating mutation generated considerable interest in the search for JAK2 inhibitors for the treatment of Philadelphia negative CMPN [11]. The current study was aimed at evaluating the status of JAK2 mutations in CMPN cases from the Western region of Saudi Arabia. Our results indicate that the percentage of JAK2 V617F exon 14 mutations in different categories of Philadelphia negative CMPN patients is similar to the previously reported frequencies elsewhere in the world [6,17]. Acquired mutations targeting exon 12 of JAK2 are reported in most JAK2 V617F-patients [18,19]. In our study, the analysis of exon 12 sequences revealed no mutations, which could be explained by the low number of patients studied. Another aspect that our study highlights is the homozygosity observed in 29% of cases with JAK2 V617F mutations. Kralovics et al, reported a significant association between homozygosity of the JAK2 V617F mutation, which occurred in approximately 27% of the patients with PV, and increased duration of disease in PV, ET, and PMF [17]. A similar observation made by others indicates that homozygosity of the mutant allele is a time-dependent clonal evolution [6]. In addition, the presence of homozygous JAK2 V617F is associated with a two-fold increase risk of thrombosis in CMPN [10]. It has been reported that the presence of homozygous
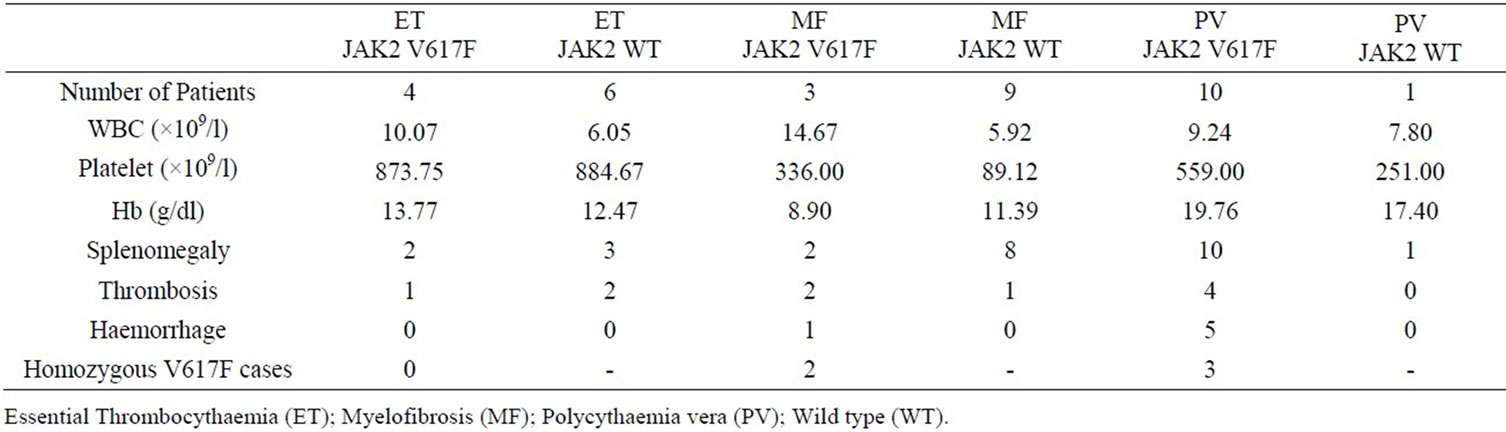
Table 2. Clinical characteristics of Ph-negative CMPN patients and their JAK2 status.
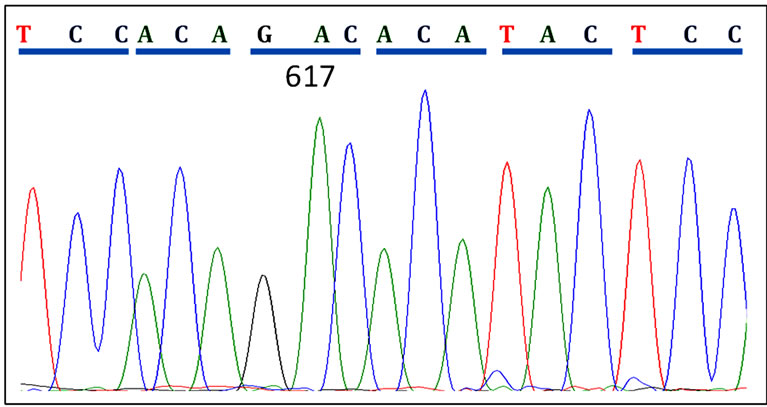 (a)
(a)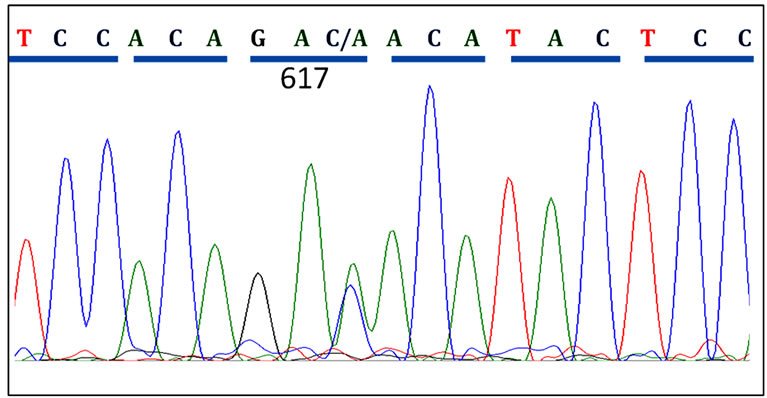 (b)
(b)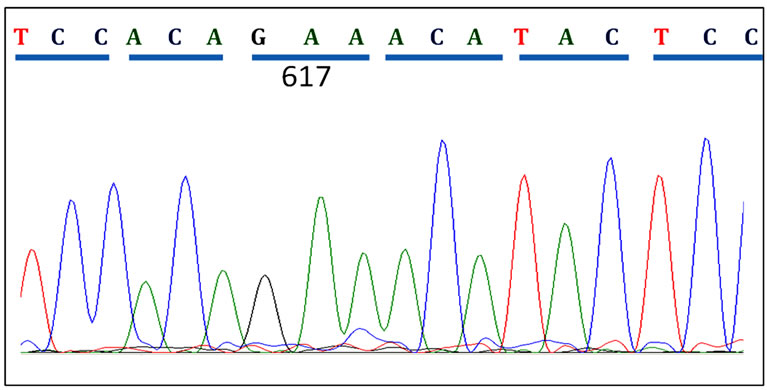 (c)
(c)
Figure 2. Patient JAK2 Exon14 sequenced using reverse primers. (a) A CMPN patient showing normal JAK2 sequence at codon 617; (b) A polycythaemia vera (PV) patient showing heterozygous mutation in codon 617; (c) Another PV patient showing homozygous mutation of the JAK2 gene at codon 617.
mutation in PV and ET patients reflect a higher leukocyte count and hemoglobin value at diagnosis, and these patients have a larger spleen volume and are also older in age. Moreover, they develop myelofibrotic transformation more than the others [20].
In conclusion, our findings strongly support the view that peripheral blood mutation screening for JAK2 V617F be introduced into the initial evaluation of patients with suspected CMPN in Saudi Arabia. An early identification of the mutation status helps in excluding a large number of secondary causes. However, since the mutation may be absent in a few cases of PV, ET and IMF, it cannot be used as a single test for making the diagnosis. Additionally, we recommend carrying this study in the whole Kingdom to include a much larger population.
5. Acknowledgements
We are really grateful to all technicians for technical assistance in hematology laboratory. This work was supported by grant from the Center of Excellence In Genomic Medicine Research, King Abdulaziz University, Ministry of Higher Education, Kingdom of Saudi Arabia
REFERENCES
- A. Tefferi and D. G. Gilliland, “Oncogenes in Myeloproliferative Disorders,” Cell Cycle, Vol. 6, No. 5, 2007, pp. 550-566. doi:10.4161/cc.6.5.3919
- C. R. Bartram, A. de Klein, A. Hagemeijer, T. van Agthoven, A. Geurts van Kessel, D. Bootsma, G. Grosveld, M. A. Ferguson-Smith, T. Davies, M. Stone, N. Heisterkamp, J. R. Stephenson and J. Groffen, “Translocation of C-Ab1 Oncogene Correlates with the Presence of a Philadelphia Chromosome in Chronic Myelocytic Leukaemia,” Nature, Vol. 306, No. 5940, 1983, pp. 277-280. doi:10.1038/306277a0
- J. D. Rowley, “A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia Identified by Quinacrine Fluorescence and Giemsa Staining,” Nature, Vol. 243, 1973, pp. 290-293. doi:10.1038/243290a0
- C. James, V. Ugo, J. P. Le Couedic, J. Staerk, F. Delhommeau, C. Lacout, L. Garcon, H. Raslova, R. Berger, A. Bennaceur-Griscelli, J. L. Villeval, S. N. Constantinescu, N. Casadevall and W. Vainchenker, “A Unique Clonal JAK2 Mutation Leading to Constitutive Signalling Causes Polycythaemia Vera,” Nature, Vol. 434, 2005, pp. 1144-1148. doi:10.1038/nature03546
- R. L. Levine, M. Loriaux, B. J. Huntly, M. L. Loh, M. Beran, E. Stoffregen, R. Berger, J. J. Clark, S. G. Willis, K. T. Nguyen, N. J. Flores, E. Estey, N. Gattermann, S. Armstrong, A. T. Look, J. D. Griffin, O. A. Bernard, M. C. Heinrich, D. G. Gilliland, B. Druker and M. W. Deininger, “The JAK2V617F Activating Mutation Occurs in Chronic Myelomonocytic Leukemia and Acute Myeloid Leukemia, But Not in Acute Lymphoblastic Leukemia or Chronic Lymphocytic Leukemia,” Blood, Vol. 106, No. 10, 2005, pp. 3377-3379. doi:10.1182/blood-2005-05-1898
- R. L. Levine, M. Wadleigh, J. Cools, B. L. Ebert, G. Wernig, B. J. Huntly, T. J. Boggon, I. Wlodarska, J. J. Clark, S. Moore, J. Adelsperger, S. Koo, J. C. Lee, S. Gabriel, T. Mercher, A. D’Andrea, S. Frohling, K. Dohner, P. Marynen, P. Vandenberghe, R. A. Mesa, A. Tefferi, J. D. Griffin, M. J. Eck, W. R. Sellers, M. Meyerson, T. R. Golub, S. J. Lee and D. G. Gilliland, “Activating Mutation in the Tyrosine Kinase JAK2 in Polycythemia Vera, Essential Thrombocythemia, and Myeloid Metaplasia with Myelofibrosis,” Cancer Cell, Vol. 7, No. 4, 2005, pp. 387-397. doi:10.1016/j.ccr.2005.03.023
- I. Dalal, E. Arpaia, H. Dadi, S. Kulkarni, J. Squire and C. M. Roifman, “Cloning and Characterization of the Human Homolog of Mouse JAK2,” Blood, Vol. 91, No. 3, 1998, pp. 844-851.
- E. Lippert, M. Boissinot, R. Kralovics, F. Girodon, I. Dobo, V. Praloran, N. Boiret-Dupre, R. C. Skoda and S. Hermouet, “The JAK2-V617F Mutation Is Frequently Present at Diagnosis in Patients with Essential Thrombocythemia and Polycythemia Vera,” Blood, Vol. 108, 2006, pp. 1865-1867. doi:10.1182/blood-2006-01-013540
- F. Passamonti, E. Rumi, D. Pietra, M. G. Della Porta, E. Boveri, C. Pascutto, L. Vanelli, L. Arcaini, S. Burcheri, L. Malcovati, M. Lazzarino and M. Cazzola, “Relation between JAK2 (V617F) Mutation Status, Granulocyte Activation, and Constitutive Mobilization of CD34+ Cells into Peripheral Blood in Myeloproliferative Disorders,” Blood, Vol. 107, 2006, pp. 3676-3682. doi:10.1182/blood-2005-09-3826
- A. Tefferi, T. L. Lasho, S. M. Schwager, J. S. Strand, M. Elliott, R. Mesa, C. Y. Li, M. Wadleigh, S. J. Lee and D. G. Gilliland, “The Clinical Phenotype of Wild-Type, Heterozygous, and Homozygous JAK2V617F in Polycythemia Vera,” Cancer, Vol. 106, No. 3, 2006, pp. 631-635. doi:10.1002/cncr.21645
- R. Tibes and R. A. Mesa, “Myeloproliferative Neoplasms 5 Years after Discovery of JAK2V617F: What Is the Impact of JAK2 Inhibitor Therapy?” Leukemia & Lymphoma, Vol. 52, No. 7, 2011, pp. 1178-1187. doi:10.3109/10428194.2011.566952
- J. W. B. R. Vardiman and N. L. Harris, “WHO Histological Classification of Chronic Myeloproliferative Diseases,” In: H. Jaffees, H. Stein and J. W. Vardiman, Eds., World Health Organization Classification of Tumors: Tumours of the Haematopoietic and Lymphoid Tissues, 2001 Edition, International Agency for Research on Cancer (IARC) Press, Lyon, France, 2001, pp. 17-44.
- World Medical Association General Assembly, “World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects,” Journal International de Bioethique International Journal of Bioethics, Vol. 15, No. 1, 2004, pp. 124-129.
- J. Chasseriau, J. Rivet, F. Bilan, J. C. Chomel, F. Guilhot, N. Bourmeyster and A. Kitzis, “Characterization of the Different BCR-ABL Transcripts with a Single Multiplex RT-PCR,” The Journal of Molecular Diagnostics, Vol. 6, 2004, pp. 343-347. doi:10.1016/S1525-1578(10)60530-2
- N. L. Harris, E. S. Jaffe, J. Diebold, G. Flandrin, H. K. Muller-Hermelink, J. Vardiman, T. A. Lister and C. D. Bloomfield, “The World Health Organization Classification of Neoplasms of the Hematopoietic and Lymphoid Tissues: Report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November, 1997,” The Hematology Journal, Vol. 1, No. 1, 2000, pp. 53-66. doi:10.1038/sj.thj.6200013
- C. James, V. Ugo, N. Casadevall, S. N. Constantinescu and W. Vainchenker, “A JAK2 Mutation in Myeloproliferative Disorders: Pathogenesis and Therapeutic and Scientific Prospects,” Trends in Molecular Medicine, Vol. 11, No. 12, 2005, pp. 546-554. doi:10.1016/j.molmed.2005.10.003
- R. Kralovics, F. Passamonti, A. S. Buser, S. S. Teo, R. Tiedt, J. R. Passweg, A. Tichelli, M. Cazzola and R. C. Skoda, “A Gain-of-Function Mutation of JAK2 in Myeloproliferative Disorders,” The New England Journal of Medicine, Vol. 352, 2005, pp. 1779-1790. doi:10.1056/NEJMoa051113
- E. Kouroupi, K. Zoi, N. Parquet, C. Zoi, J. J. Kiladjian, V. Grigoraki, W. Vainchenker, F. Lellouche, C. Marzac, M. H. Schlageter, C. Dosquet, L. M. Scott, P. Fenaux, D. Loukopoulos, C. Chomienne and B. Cassinat, “Mutations in Exon 12 of JAK2 Are Mainly Found in JAK2 V617F-Negative Polycythaemia Vera Patients,” British Journal of Haematology, Vol. 142, 2008, pp. 676-679. doi:10.1111/j.1365-2141.2008.07223.x
- L. M. Scott, W. Tong, R. L. Levine, M. A. Scott, P. A. Beer, M. R. Stratton, P. A. Futreal, W. N. Erber, M. F. McMullin, C. N. Harrison, A. J. Warren, D. G. Gilliland, H. F. Lodish, A. R. Green, “JAK2 Exon 12 Mutations in Polycythemia Vera and Idiopathic Erythrocytosis,” The New England Journal of Medicine, Vol. 356, 2007, pp. 459-468. doi:10.1056/NEJMoa065202
- A. M. Vannucchi, E. Antonioli, P. Guglielmelli, A. Rambaldi, G. Barosi, R. Marchioli, et al., “Clinical Profile of Homozygous JAKV2 V617F Mutation in Patients with Polycythemia Vera or Essential Thrombocythemia,” Blood, Vol. 110, 2007, pp. 840-846. doi:10.1182/blood-2006-12-064287
NOTES
*Corresponding author.

