Open Journal of Medicinal Chemistry
Vol.06 No.03(2016), Article ID:68568,8 pages
10.4236/ojmc.2016.63004
Evaluation of Benzamide Derivatives as New Influenza A Nucleoprotein Inhibitors
Jinxi Liao1, Huimin Cheng2, Junting Wan1, Panyu Chen1, Yingjun Li2,3, Ke Ding2, Micky D. Tortorella1, Zhengchao Tu1*, Yanmei Zhang1*
1Drug Discovery Pipeline, Guangzhou Institutes of Biomedicine and Health, Science City, Guangzhou, China
2Institute of Chemical Biology, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou, China
3University of Chinese Academy of Sciences, Beijing, China

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 28 April 2016; accepted 15 July 2016; published 18 July 2016
ABSTRACT
Virus nucleoprotein (NP) is an emerging target for drug development for Influenza. We designed benzamide derivatives as new inhibitors of NP that demonstrate good potency in blocking influenza A. Screening revealed that compound 39 was the most potent molecule in the series, exhibiting IC50 values of 0.46 and 0.27 μM in blocking the replication of H3N2 (A/HK/8/68) and (A/WSN/33) influenza A viral strains. The observed inhibition of viral replication correlated well with cytopathic protection. Furthermore, based on computational analysis and fluorescence microscopy, it was determined that compound 39 inhibited nuclear accumulation by targeting influenza A viral nucleoproteins. Finally, the rodent pharmacokinetic profile of compound 32 displayed half-life of greater than 4 hours and bioavailability greater than 20%, suggesting this class of molecules had drug-like properties.
Keywords:
Benzamides, Nucleoprotein, Anti-Influenza, NP Inhibitors, Nucleozin

1. Introduction
Influenza is a contagious and infectious respiratory disease mediated by the flu virus. Every year the flu makes its way across the world causing illness and even death, particularly in the elderly and young who are more likely to be immunocompromised [1] . As an example of the lethality of this negative-sense single stranded RNA virus, it is estimated that in 2009, approximately 16,000 people died in Mexico as a result of the swine flu, demonstrating the virulence of this class of virus. Influenza viruses are categorized into three serological subtypes, including type A, B and C [2] . Influenza type A is the most virulent of the three and is the primary focus of vaccination. Influenza A viruses contain 8 segmented [3] RNA sections and are classified based on NA and HA types that include 9 NA subtypes (N1 - N9) and 16 HA subtypes (H1 - H16). Influenza A viruses undergo constant mutation and reassembly of its genetic material resulting in evasion of human immunity and therapies [4] . Although rare, this viral genetic shift can even result in interspecies transmission from animals to humans. Case in point is the reassembly of the avian influenza A (H7N9) virus [5] originating in China in 2013, resulting in the infection of at least 134 people and causing 35 reported deaths.
Influenza is currently treated by two classes of drugs including small molecules and vaccines. Among the small molecules are M2 ion channel blockers such as Amantidine (Symmetrel) and Rimantidine (Flumadine) and neuraminidase inhibitors such as Oseltamivir (Tamiflu) and Zanamivir (Relenza) [6] . Unfortunately, the effectiveness of these drugs has been compromised by the emergence of drug-resistant viral mutations [7] - [9] . For example, since 2005 due to a H274Y mutation in neuraminidase, Oseltamivir is now relatively ineffective against the flu [10] - [12] and all isolates of the H3N2 viruses are now resistant to the M2 blockers [13] [14] . The speed and rate of mutation by the influenza virus highlights the need for developing new medicines targeting different mechanisms and viral proteins.
One such protein that has garnished attention is the influenza virus A nucleoprotein (NP) coded by the fifth genome segment. NP is expressed at very high levels throughout the course of infection and has several critical functions [15] [16] such as genome trafficking, vRNA synthesis and viral assembly. Unlike other targets, like hemagglutinin (HA) and neuraminidase (NA), NP is relatively conserved in the influenza virus genome making this protein an attractive target for drug development [17] - [21] .
The molecule nucleozin (Figure 1) was shown to be a potent blocker of NP accumulation in the nucleus, reported by Yuen et al. [22] and Wong et al. [23] in 2010. It was determined that nucleozin and related analogs catalyzed the aggregation of nucleoprotein, thus preventing viral nuclear accumulation resulting in potent anti-influenza activity. In 2011, the discovery and optimization of nucleozin like molecules by Samuel [24] et al. were shown to have even greater efficacy in blocking influenza and co-crystallization with one of the molecules with NP showed an unusual NP_A: NP_B dimeric subunit bearing two protein bridging ligands in an antiparallel arrangement. This unusual quaternary complex suggested that the NP inhibitors were inducing the formation of stable NP oligomers, blocking viral nuclear accumulation. In 2012, our team [25] developed new potent NP inhibitors by employing bio-isosteric replacement strategies and scaffold-hopping, but unfortunately the molecules displayed poor pharmacokinetics in rodents due to metabolic instability thereby limiting their utility as drugs. The focus of this paper is on chemical improvements that we make to the benzamide based NP inhibitors.
Figure 1. Chemical structures of nucleoprotein inhibitors.
2. Results and Discussion
2.1. Chemistry
Based on modelling we assumed the isoxazole moiety of nucleozin is a factor for the observed metabolic instability for this class of molecules [24] , so as a first strep we replaced the isoxazole motif of nucleozin into a simple phenyl group and evaluated the influence on stability and potency against influenza A. Moreover, a series of benzamide derivatives were made with the hopes of further improving both the potency and pharmacokinetics of the drugs.
Benzamide based derivatives were prepared by direct condensation using substituted benzoic acids with 1-(2- chloro-4-nitrophenyl) piperazine in the presence of EDCI and Et3N at room temperature (Scheme 1). 1-(2-chloro- 4-nitrophenyl) piperazine was made using previously described methods and the substituted benzoic acids were made using commercial chemicals as starting points for modification. According to our previous publication [25] , the 2-chloro-4-nitroaniline (ring B) was found to be important for anti-influenza potency. So, in this study our modifications were primarily focused on the A ring where multiple substituents were introduced to evaluate their effect on activity.
2.2. MDCK Cell-Based CPE Assays
Using Madin-Darby canine kidney (MDCK) cells, compounds were tested for their potency in blocking theinfluenza A viral mediated, cytopathic effect (CPE), summarized in Table 1. Nucleozin (compound 3), which was used as positive control, showed good inhibition of the replication of H3N2 (A/HK/8/68) and H1N1 (A/WSN/33) strains with IC50 values of 1.93 and 1.15 μM, respectively.
As hoped, substituted groups in phenyl A, significantly improved the anti-influenza activity. Based on this approach we substituted a methyl, chloro, bromo, or nitro group into the ortho-, meta- or para-position of the A ring (compound 11 to 22) and found that the ortho- or para-substitution (compound 11, 13, 14, 16, 17, 19, 20 and 22) increased potency while the meta-substitution (compound 12, 15, 18 and 21) had only a modest effect on activity. Case in point, when we added a nitro group into the para-position of the A ring (compound 22), a dramatic increase in potency was observed with IC50 values of 3.45 and 3.01 μM for blocking the replication of H3N2 (A/HK/8/68) and H1N1 (A/WSN/33), respectively. This is about six times more potent than compound 10. Other substitutions at the ortho- or para-position, other than compound 29 showed an increase in potency demonstrating a clear structure activity relationship.
Di-substituted derivatives with 2-, 6- and 2-, 4-substitutions in the phenyl A ring (compound 30 to 38) also showed better efficacy compared to the mono substituted analogs. For example, compound 34 displayed IC50 values of 1.78 and 0.52 μM against H3N2 (A/HK/8/68) and H1N1 (A/WSN/33). Compound 39 was made by introducing a third substituent into the 4-position of the phenyl A ring of compound 33, and this tri-substituent phenyl A ring displayed even better efficacy in blocking the replication of H3N2 (A/HK/8/68) and H1N1 (A/WSN/33) with IC50 values of 0.46 and 0. 27 μM.
The safety of the compounds was tested for potential cytotoxic effects using MDCK cells, results also summarized in Table 1. Most of the compounds showed no cellular growth inhibition against MDCK cells up to 100 μM, although our most potent antiviral derivative, compound 39, displayed some toxicity against MDCK cells with an IC50 value of 1.53 μM.
2.3. Plaque Reduction Assay
The plaque reduction assay was used to further evaluate the efficacy of lead molecules 33 and 39. As shown in Figure 2, compound 33 and 39 dose dependently blocked plaque formation in MDCK cells following A/WSN/33 (H1N1) influenza viral infection.
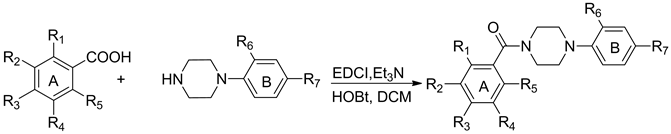
Scheme 1. Synthesis of Compounds 10 to 39. Reagents and conditions: EDCI, Et3N, room temperature, 8.0 h, 42.4% - 88.5%.
Table 1. Inhibitory activity of the compounds against influenza a replication
aInfluenza A/HK/8/68 (H3N2) stains. bInfluenza A/WSN/33 (H1N1) stains. cAntiproliferation against MDCK cells. The data are means of results from at least three independent experiments.
The plaque formation was inhibited at concentrations as low as 0.11 μM while complete inhibition was seen at concentrations of ~1 μM for both compounds. These results were in alignment with the CPE assay.
In order to better understand the mechanism by which nucleozin triggers the aggregation of NP and blocking of its nuclear accumulation in influenza infected MDCK cells, the effects of compound 39 was evaluated using fluorescence microscopy (Figure 3). The data showed that molecule 39 blocked nuclear accumulation of the influenza virus A NP, following infection with H1N1 (A/WSN/33), trapping the protein in the cytoplasm of the MDCK cells. These data suggest that the nucleoproteins are the target of compound 39.
Figure 2. Compound 33 and 39 inhibited plaque formation in influenza virus infected MDCK cells. Monolayer MDCK cells were infected with A/WSN/33 (H1N1) at 0.01 MOI (multiplicity of infection) and overlaid with 1% agar containing various concentrations of 33 (upper panel), 39 (middle panel) and Nucleozin (bottom panel). At 72 h post infection, plaques were visualized by staining with crystal violet. The results shown are representative of at least three independent experiments.
Figure 3. Compound 39 blocked nucleoproteins of influenza virus A into the nucleus of MDCK cells that were infected with A/WSN/33 (H1N1) virus. Multiplicity of infection (MOI) of 10 in the absence or presence of 10 μM Nucleozin (middle row) or 39 (bottom row). At 4.0 h postinfection, cells were fixed and stained with DAPI for nucleus (blue), and FITC-tagged anti-influenza A NP antibodies specific for viral nucleoprotein (green), respectively. Images were visualized by confocal microscopy. The illustration is representative of experiments in triplicate.
Figure 4. Compound 39 (blue) binds to nucleoproteins in a similar manner to that of the nucleozin (pink). (a) 39-NP complex structure predicted by molecular docking. (b) Overlay of 39 and nucleozin in the nucleoprotein dimer interface pocket.
Table 2. Pharmacokinetic profile of compound 32 in ratsa.
aSD rats (male, 4 animals per group) weighted 180 - 220 g were used for the study.
2.4. Docking Study
Computational docking analysis predicts that compound 39 binds to the nucleoproteins in a similar manner asnucleozin and related analogues (Figure 4). The carbonyl group of compound 39 forms a hydrogen bond to the hydroxyl group of serine residue 376 in NP_A and the chloro group in ring B forms a halogen bond with tyrosine residue 52 in NP_B. In addition, tyrosine 289 in NP_B interacts with the 2-chloro-4-nitro-phenyl moiety of 39, forming a strong π-π stacking interaction.
2.5. PK Study
As mentioned before, poor pharmacokinetic properties limit nucleozin and related derivatives as drug candidates, thus we investigate the pharmacokinetic properties of our new benzamide derivatives as potentially better drug like molecules. As a representative molecule, compound 32 was dosed orally in rats at 25 mg/kg or by injection at 5 mg/kg and displayed a promising pharmacokinetic profile summarized in Table 2. The half-life was 4.6 h and showed reasonable oral bioavailability of ~21%, suggesting that these benzamide based NP inhibitors may be viable drug which leads to further optimization and development.
In summary, we have successfully designed and synthesized a new series of benzamide derivatives with potent anti-influenza A activity. Unlike the first generation of NP inhibitors such as nucleozin and related derivatives, this series offers acceptable pharmacokinetic properties differentiating this class as a drug like that warrants further preclinical development.
Acknowledgements
The research work was financially supported by the Guangzhou Science & Technology Project (2011Y2-00026 and 201508020131) to support GIBH Drug Discovery Pipeline Development (Y. Z.), the project funded by National Science Foundation of China (21202169) (Y. Z.), CAS Key Technology Talent Program China (2013) (Z. T.) and Guangdong Science and Technology Plan projects, China (Grant No. 2013B040200031) (Z. T.).
Cite this paper
Jinxi Liao,Huimin Cheng,Junting Wan,Panyu Chen,Yingjun Li,Ke Ding,Micky D. Tortorella,Zhengchao Tu,Yanmei Zhang,1 1, (2016) Evaluation of Benzamide Derivatives as New Influenza A Nucleoprotein Inhibitors. Open Journal of Medicinal Chemistry,06,43-50. doi: 10.4236/ojmc.2016.63004
References
- 1. Shen, Y.-F., Chen, Y.-H., Chu, S.-Y., Lin, M.-I., Hsu, H.-T., Wu, P.-Y., Wu, C.-J., Liu, H.-W., Lin, F.-Y., Lin, G., Hsu, P.-H., Yang, A.-S., Cheng, Y.-S.E., Wu, Y.-T., Wong, C.-H. and Tsai, M.-D. (2011) E339 ... R416 Salt Bridge of Nucleoprotein as a Feasible Target for Influenza Virus Inhibitors. Proceedings of the National Academy of Sciences of the United States of America, 108, 16515-16520.
http://dx.doi.org/10.1073/pnas.1113107108 - 2. Lejal, N., Tarus, B., Bouguyon, E., Chenavas, S., Bertho, N., Delmas, B., Ruigrok, R.W.H., Di Primo, C. and Slama-Schwok, A. (2013) Structure-Based Discovery of the Novel Antiviral Properties of Naproxen against the Nucleoprotein of Influenza A Virus. Antimicrobial Agents and Chemotherapy, 57, 2231-2242.
http://dx.doi.org/10.1128/AAC.02335-12 - 3. Kao, R.Y., Yang, D., Lau, L.-S., Tsui, W.H.W., Hu, L., Dai, J., Chan, M.-P., Chan, C.-M., Wang, P., Zheng, B.-J., Sun, J., Huang, J.-D., Madar, J., Chen, G., Chen, H., Guan, Y. and Yuen, K.-Y. (2010) Identification of Influenza A Nucleoprotein as an Antiviral Target. Nature Biotechnology, 28, 600-605.
http://dx.doi.org/10.1038/nbt.1638 - 4. Su, C.-Y., Cheng, T.-J.R., Lin, M.-I., Wang, S.-Y., Huang, W.-I., Lin-Chu, S.-Y., Chen, Y.-H., Wu, C.-Y., Lai, M.M.C., Cheng, W.-C., Wu, Y.-T., Tsai, M.-D., Cheng, Y.-S.E. and Wong, C.-H. (2010) High-Throughput Identification of Compounds Targeting Influenza RNA-Dependent RNA Polymerase Activity. Proceedings of the National Academy of Sciences of the United States of America, 107, 19151-19156.
http://dx.doi.org/10.1073/pnas.1013592107 - 5. Gerritz, S.W., Cianci, C., Kim, S., Pearce, B.C., Deminie, C., Discotto, L., McAuliffe, B., Minassian, B.F., Shi, S., Zhu, S., Zhai, W., Pendri, A., Li, G., Poss, M.A., Edavettal, S., McDonnell, P.A., Lewis, H.A., Maskos, K., Moertl, M., Kiefersauer, R., Steinbacher, S., Baldwin, E.T., Metzler, W., Bryson, J., Healy, M.D., Philip, T., Zoeckler, M., Schartman, R., Sinz, M., Leyva-Grado, V.H., Hoffmann, H.-H., Langley, D.R., Meanwell, N.A. and Krystal, M. (2011) Inhibition of Influenza Virus Replication via Small Molecules That Induce the Formation of Higher-Order Nucleoprotein Oligomers. Proceedings of the National Academy of Sciences of the United States of America, 108, 15366-15371.
http://dx.doi.org/10.1073/pnas.1107906108 - 6. Cheng, H., Wan, J., Lin, M.-I., Liu, Y., Lu, X., Liu, J., Xu, Y., Chen, J., Tu, Z., Cheng, Y.-S.E. and Ding, K. (2012) Design, Synthesis, and in Vitro Biological Evaluation of 1 H-1, 2, 3-Triazole-4-carboxamide Derivatives as New Anti-influenza A Agents Targeting Virus Nucleoprotein. Journal of Medicinal Chemistry, 55, 2144-2153.
http://dx.doi.org/10.1021/jm2013503 - 7. Fedichev, P., Timakhov, R., Pyrkov, T., Getmantsev, E. and Vinnik, A. (2011) Structure-Based Drug Design of a New Chemical Class of Small Molecules Active against Influenza A Nucleoprotein in Vitro and in Vivo. PLoS Currents, 3, RRN1253.
http://dx.doi.org/10.1371/currents.rrn1253 - 8. Hagiwara, K., Kondoh, Y., Ueda, A., Yamada, K., Goto, H., Watanabe, T., Nakata, T., Osada, H. and Aida Y. (2010) Discovery of Novel Antiviral Agents Directed Against the Influenza A Virus Nucleoprotein Using Photo-Cross-Linked Chemical Arrays. Biochemical and Biophysical Research Communications, 394, 721-727.
http://dx.doi.org/10.1016/j.bbrc.2010.03.058 - 9. Gong, J.Z., Fang, H., Li, M.Y., Liu, Y., Yang, K.H., Liu, Y.Z. and Xu, W.F. (2009) Potential Targets and Their Relevant Inhibitors in Anti-Influenza Fields. Current Medicinal Chemistry, 16, 3716-3739.
http://dx.doi.org/10.2174/092986709789104984 - 10. Portela, A. and Digard, P. (2002) The Influenza Virus Nucleoprotein: A Multifunctional RNA-Binding Protein Pivotal to Virus Replication. Journal of General Virology, 83, 723-734.
http://dx.doi.org/10.1099/0022-1317-83-4-723 - 11. Huang, T.S., Palese, P. and Krystal, M. (1990) Determination of Influenza Virus Proteins Required for Genome Replication. Journal of Virology, 64, 5669.
- 12. Layne, S.P., Monto, A.S. and Taubenberger, J.K. (2009) Pandemic Influenza: An Inconvenient Mutation. Science, 323, 1560-1561.
http://dx.doi.org/10.1126/science.323.5921.1560 - 13. Regoes, R.R. and Bonhoeffer, S. (2006) Emergence of Drug-Resistant Influenza Virus: Population Dynamical Considerations 2009 Pandemic Influenza: An Inconvenient Mutation. Science, 312, 389-391.
http://dx.doi.org/10.1126/science.1122947 - 14. Mitrasinovic, P.M. (2010) Advances in the Structure-Based Design of the Influenza A Neuraminidase Inhibitors. Current Drug Targets, 11, 315-326.
http://dx.doi.org/10.2174/138945010790711932 - 15. Ives, J.A.L., Carr, J.A., Mendel, D.B., Tai, C.Y., Lambkin, R., Kelly, L., Oxford, J.S., Hayden, F.G. and Roberts, N.A. (2002) The H274Y Mutation in the Influenza A/H1N1 Neuraminidase Active Site Following Oseltamivir Phosphate Treatment Leave Virus Severely Compromised both in Vitro and in Vivo. Antiviral Research, 55, 307-317.
http://dx.doi.org/10.1016/S0166-3542(02)00053-0 - 16. Carr, J., Ives, J., Kelly, L., Lambkin, R., Oxford, J., Mendel, D., Tai, L. and Roberts, N. (2002) Influenza Virus Carrying Neuraminidase with Reduced Sensitivity to Oseltamivir Carboxylate Has Altered Properties in Vitro and Is Compromised for Infectivity and Replicative Ability in Vivo. Antiviral Research, 54, 79-88.
http://dx.doi.org/10.1016/S0166-3542(01)00215-7 - 17. Gubareva, L., Okomo-Adhiambo, M., Deyde, V., Sheu, T.G., Garten, R., Smith, C., Barnes, J., Myrick, A., Hillman, M., Shaw, M., Bridges, C., Klimov, A. and Cox, N. (2009) Update: Drug Susceptibility of Swine-Origin. JAMA- Journal of the American Medical Association, 301, 2086.
- 18. Deyde, V.M., Xu, X.Y., Bright, R.A., Shaw, M., Smith, C.B., Zhang, Y., Shu, Y.L., Gubareva, L.V., Cox, N.J. and Klimov, A.I. (2007) Surveillance of Resistance to Adamantanes among Influenza A (H3N2) and A (H1N1) Viruses Isolated Worldwide. The Journal of Infectious Diseases, 196, 249-257.
http://dx.doi.org/10.1086/518936 - 19. Bright, R.A., Shay, D.K., Shu, B., Cox, N.J. and Klimov, A.I. (2006) Adamantane Resistance among Influenza A Viruses Isolated Early during the 2005-2006 Influenza Season in the United States. AMA-Journal of the American Medical Association, 295, 891-894.
http://dx.doi.org/10.1001/jama.295.8.joc60020 - 20. Du, J., Cross, T.A. and Zhou,H.X. (2012) Recent Progress in Structure-based Anti-influenza Drug Design. Drug Discovery Today, 17, 1111-1120.
http://dx.doi.org/10.1016/j.drudis.2012.06.002 - 21. Gao, R.B., Cao, B., Hu, Y.W., Feng, Z.J., Wang, D.Y., Hu, W.F., Chen, J., Jie, Z.J., Qiu, H.B., Xu, K., Xu, X.W., Lu, H.Z., Zhu, W.F., Gao, Z.C., Xiang, N.J., Shen, Y.Z., He, B.Z., Gu, Y., Zhang, Z.Y., Yang, Y., Zhao, X., Zhou, L., Li, X.D., Zou, S.M., Zhang, Y., Li, X.Y., Yang, L., Guo, J.F., Dong, J., Li, Q., Dong, L.B., Zhu, Y., Bai, T., Wang, S.W., Hao, P., Yang, W.Z., Zhang, Y.P., Han, J., Yu, H.J., Li, D.X., Gao, G.F., Wu, G.Z., Wang, Y., Yuan, Z.H. and Shu, Y.L. (2013) Human Infection with a Novel Avian-origin Influenza A (H7N9) Virus. The New England Journal of Medicine, 368, 1888-1897.
http://dx.doi.org/10.1056/NEJMoa1304459 - 22. Wilson, J.C. and von Itzstein, M. (2003) Recent Strategies in the Search for New Anti-Influenza Therapies. Current Drug Targets, 4, 389-408.
http://dx.doi.org/10.2174/1389450033491019 - 23. Ghedin, E., Sengamalay, N.A., Shumway, M., Zaborsky, J., Feldblyum, T., Subbu, V., Spiro, D.J., Sitz, J., Koo, H., Bolotov, P., Dernovoy, D., Tatusova, T., Bao, Y.M., St George, K., Taylor, J., Lipman, D.J., Fraser, C.M., Taubenberger, J.K. and Salzberg, S.L. (2005) Large-Scale Sequencing of Human Influenza Reveals the Dynamic Nature of Viral Genome Evolution. Nature, 437, 1162-1166.
http://dx.doi.org/10.1038/nature04239 - 24. Palese, P. and Young, J.F. (2010) Structural Basis of Preexisting Immunity to the 2009 H1n1 Pandemic Influenza Virus. Science, 215, 1468-1474.
http://dx.doi.org/10.1126/science.7038875 - 25. Cox, N.J. and Subbarao, K. (2000) Global Epidemiology of Influenza: Past and Present. Annual Review of Medicine, 51, 407-421.
http://dx.doi.org/10.1146/annurev.med.51.1.407
NOTES
*Corresponding authors.







