Computational Chemistry
Vol.03 No.04(2015), Article ID:60843,8 pages
10.4236/cc.2015.34006
2D-QSAR Study of a Series of Pyrazoline-Based Anti-Tubercular Agents Using Genetic Function Approximation
Hemal M. Soni1, Popatbhai K. Patel2, Mahesh T. Chhabria3, Dharmraj N. Rana4, Bhushan M. Mahajan1, Pathik S. Brahmkshatriya4*
1M/S Piramal Enterprises Ltd., Piramal Enterprises Limited-Discovery Solutions, Plot No. 18, Pharmaceutical Special Econonic Zone, Village Matoda, Ta. Sanand, Ahmedabad, India
2M.G. Science Institute, Dada Saheb Mavlankar Campus, Opp. Gujarat University, Ahmedabad, India
3Department of Pharmaceutical Chemistry, L. M. College of Pharmacy, Ahmedabad, India
4Oxygen Healthcare Res. Pvt. Ltd., Plot No. 35, Panchratna Industrial Estate, Near IBP Laxminarayan Petrol Pump, Changodar, Sarkhej-Bavla Road, Ahmedabad, India
Email: *pathik26@yahoo.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 11 October 2015; accepted 27 October 2015; published 30 October 2015
ABSTRACT
A series of pyrazoline-based new heterocycles have recently been synthesized from our group where some of the compounds display potent anti-tubercular activity against Mycobacterium tuberculosis H37Rv. In order to further explore the potency of the compounds, quantitative structure activity relationship study is carried out using genetic function approximation. Statistically significant (r2 = 0.85) and predictive 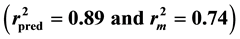 QSAR models are developed. It is evident from the QSAR study that majority of the anti-tubercular activity is found to be driven by lipophilicity. Also, molecular solubility, Jurs and shadow descriptors influence the biological activity significantly. Also, positive contribution of molecular shadow descriptors suggests that molecules with bulkier substituents are more likely to enhance anti-tubercular activity. Since the developed QSAR models are found to be statistically significant and predictive, they potentially can be applied for predicting anti-tubercular activity of new molecules for prioritization of molecules for synthesis.
QSAR models are developed. It is evident from the QSAR study that majority of the anti-tubercular activity is found to be driven by lipophilicity. Also, molecular solubility, Jurs and shadow descriptors influence the biological activity significantly. Also, positive contribution of molecular shadow descriptors suggests that molecules with bulkier substituents are more likely to enhance anti-tubercular activity. Since the developed QSAR models are found to be statistically significant and predictive, they potentially can be applied for predicting anti-tubercular activity of new molecules for prioritization of molecules for synthesis.
Keywords:
QSAR, Genetic Function Approximation, Descriptors, Cross-Validation

1. Introduction
World Health Organization (WHO) estimates that almost one-third of the world’s population, (~2 billion people) is infected with the tuberculosis [1] . Every year, more than 8 million people develop an active form of this disease, which claims the lives of nearly 2 million. WHO estimated in 2002 that if the worldwide spread of tuberculosis was left unchecked, it would be responsible for nearly 36 million more deaths by 2020. Effective and specific anti-tubercular drugs are still not found and classical antibiotics are currently being used for curing tuberculosis. However, effectiveness of such treatment is rather controversial [2] . Multidrug-resistant TB (MDR- TB), a form of TB that does not respond to the first-line TB drugs and extensively drug-resistant TB (XDR-TB), an MDR-TB with resistance to aminoglycosides and fluoroquinolones has become a serious threat to control and treatment of tuberculosis. There are also a few cases reported of totally drug resistant tuberculosis (TDR-TB); which has raised alarming concerns on the existing drug regimen [3] . This implies urgent need to discover newer anti-tubercular agents with newer molecular mechanisms.
Quantitative structure activity relationship (QSAR) is one of the most widely used tools to design newer candidates for several therapeutic areas [4] - [6] . It provides useful insights into the structural features which are responsible for the biological activity and help to generate a mathematical model which can predict activity of untested compounds quantitatively. QSAR study usually leads to a predictive formula by correlation of physicochemical properties of a congeneric series with the biological activity [7] .
Earlier from our laboratory, a series of pyrazoline-based benzoxazoles are identified as potent anti-tubercu- lar agents [8] [9] . In order to further investigate the potency of the molecule as a part of lead optimization program, we carry out QSAR study by using Genetic Function Approximation (GFA) technique [10] . GFA algorithm is a novel approach to create structure―activity models. It searches QSAR models automatically by combining statistical modeling with genetic algorithm tools. Typically, thousands of candidate models are generated and tested during evolution. However, only the superior (best) models survive; which are used as “parents” to create the next generation of candidate models. Previously, we have successfully applied GFA to generate a variety of QSAR models [4] [5] . Such models provide useful structure-activity insights, which can be used for prioritization of synthetic efforts to generate and lead optimization strategies.
2. Experimental
2.1. Data Set
In present studies, a series of substituted pyrazoline-based compounds reported by Rana et al. as potent anti-tu- bercular agents was selected [8] [9] . Fifty four compounds were randomly divided into training and test set, the former set consisting of thirty nine compounds and the remaining fifteen compounds were taken as the test set. Structures of all the compounds used for 2D-QSAR analysis and their anti-tubercular activity (MIC, µg/mL) are given in Table 1. For all the compounds, the experimental values of biological activity (MIC) are used in the negative logarithmic scale (pMIC) to achieve normal distribution. Structures of all compounds were sketched by using visualizer module of Discovery Studio 2.1 software (Accelrys Inc., USA). CHARMM force field was used for the calculation of potential energy. Energy minimization of all the compounds was done using Smart Minimizer method until the root mean square (RMS) gradient value becomes smaller than 0.001 kcal/mol Å. This was followed by geometry optimization by semi empirical MOPAC-AM1 method (Astin Method-1).
2.2. Descriptor Calculation
“Calculate Molecular Properties” protocol of the Discovery Studio 2.1 was used to calculate various physicochemical descriptors like structural, thermodynamic, steric, electronic and quantum mechanical descriptors. Further, a correlation matrix of the molecular descriptors was generated and highly correlated descriptors with a correlation value of 0.6 or above were discarded from the study. Remaining least correlated descriptors were used to develop 2D-QSAR models. Descriptors included in developing 2D-QSAR models are listed and described in Table 2.
2.3. Regression Analysis
The advantage of GFA is that the data set is being modeled to generate a population of equations rather than one
Table 1. Chemical structures and biological activity of training set (1-39) and test set (40-54) compounds.

Table 2. List of descriptors used in the study.
single equation for descriptor-activity correlation. GFA is genetic principle based method of variable selection, which combines Holland’s genetic algorithm and Friedman’s multivariate adaptive regression splines. Thus, it evolves the population of equations that best fit the training set data.
In GFA, a particular number of equations (set at 100 by default) are randomly generated. The pairs of “parent” equations then are chosen randomly from this set of 100 equations. After this, “crossover” operations are performed at random. The number of crossing over was set at 5000 by default. The goodness of each progeny equation is assessed by Friedman’s lack of fit (LOF) score

where c is the number of basis functions in the model, LSE is the least-squares error, p is the number of descriptors, d is smoothing parameter, and m is the number of observations in the training set. The smoothing parameter controls the scoring bias between equations of different sizes. It was set at default value of 0.5. GFA crossover of 5000 was set to give reasonable convergence. The length of equation was fixed to six terms, the population size was established as 100, and the mutation probability was specified as 0.1. Best three equations, out of the 100 equations, were chosen based on the statistical parameters like LOF, regression coefficient (r), adjusted regression coefficient (radj), cross-validated regression coefficient (rcv) and F-test values.
2.4. Validation Test
Variance inflation factor (VIF) analysis was performed to check the inter-correlation of descriptors. VIF value is calculated from 1/1 − r2, where r2 is the multiple correlation coefficient of one molecular descriptor’s effect regressed on the remaining descriptors. VIF value greater than10 suggests chance-correlation and hide the information of molecular descriptors by inter-correlation of descriptors [11] .
It is proven that a high value of statistical characteristics r and F and low value of s and LOF need not be the criteria of a highly predictive model. Thus, in order to evaluate the predictive ability of the 2D-QSAR model, the external predictability method described by Roy et al. was used [12] . It was determined by calculating the value of predictive r2 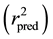 using the following equation
using the following equation
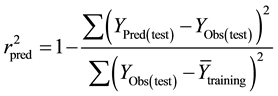
where, YObs(test) and YPred(test) are the observed and predicted activity values, respectively, of the test set compounds and Ytraining is the mean activity value of the training set.
3. Results and Discussion
In the present study, 31 descriptors were selected initially for correlation with anti-tubercular activity. The 31 preselected descriptors represented different class of descriptors such as quantum mechanical, steric, geometric, thermodynamic, and electronic. The descriptors were correlated with training set using GFA methodology. Initially, 100 2D-QSAR equations with six descriptors were generated. The results of the best three models are given in Table 3 along with their regression statistics.
For a statistically significant model, it is inevitable that the descriptors evolved in the equation should be least inter-correlated with each other. In the present study, the inter-correlation of the descriptors used in the selected models was found to be very low. The correlation matrix for the used descriptors is shown in Table 4.
Further to check the inter-correlation of descriptors, variance inflation factor (VIF) analysis was performed (as described in Section 2.4). VIF values of these descriptors were found to be 2.010 (ALogP), 1.243 (Jurs_RNCG), 2.558 (Apol), 1.366 (Jurs_DPSA_1), 1.520 (Shadow_XZ) and 1.585(Molecular_Solubility). All the VIF values were found to be less than 10, which suggest very less multi-collinearity within descriptors. The models were also evaluated for their predictive power, i.e. internal and external cross-validation. The results for Equation (1) are summarized in Table 5 and Table 6.
Figure 1 and Figure 2 show the plot of observed Vs predicted activity for training and test set compounds, respectively as per Equation (1). It was seen that the models displayed 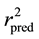 and
and 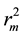 in the acceptable range [12] .
in the acceptable range [12] .
The descriptors used in the study were found to have significant influence on the biological activity as seen
Table 3. Selected 2D-QSAR equations and their regression statistics.
Table 4. Correlation matrix of the descriptors used in the equations.
Figure 1. Plot of observed Vs predicted pMIC values of training set compounds (as per Equation (1)).
from their high coefficients values. Noticeably, the activity was found to be governed chiefly through lipophilicity (AlogP). As seen from the positive coefficient, lipophilicity positively influenced the activity. Indeed, com- pounds with halogens (bromo/chloro, 2, 7, 16, 20) were found to possess high anti-tubercular activity whereas compounds with polar groups (9-15) were found to be less active. Jurs descriptors are a group of molecular descriptors which combine electronic and shape information to characterize molecules [13] . They are calculated by mapping atomic partial charges on solvent-accessible surface areas of individual atoms. Jurs_RNCG is charge of most negative atom divided by the total negative charge. Jurs_DPSA_1 is partial positive solvent-accessible surface area minus partial negative solvent-accessible surface area. A critical analysis of the generated equations
Table 5. Observed and predicted pMIC values of training set compounds (as per Equation (1)).
Figure 2. Plot of observed Vs predicted pMIC values of test set compounds (as per Equation (1)).
Table 6. Observed and predicted pMIC values of test set compounds (as per Equation (1)).
suggested negative contribution of these descriptors on biological activity. This means that the charge distribution within the molecules serves as the driving force for intermolecular interactions and the higher the relative charge the smaller the interactions. The above fact is exemplified from compounds 2, 20, 30 where lower values of the above descriptors resulted in increase in activity. Another set of geometrical descriptors, Molecular Shadow descriptors like Shadow_XZ (area of the molecular shadow in the XZ plane) also showed significant contribution to anti-tubercular activity with the coefficient being positive. This shows that molecules with bulkier substituents (2, 20, 30, 35) are more likely to show activity. In consistent with the above correlation, compounds 1, 5, 13 and 21 (with one or more H substituents) stood out as less active due to low values of Shadow_XZ. Apol (the sum of the atomic polarizabilities) also contributed positively to anti-tubercular activity. However, its low co-efficient signals its low contribution as compared to the other descriptors.
4. Conclusion
Developed 2D-QSAR models were found to be statistically significant as seen from their regression statistics. Also, during internal and external cross-validation studies, very low residuals were obtained which suggested that developed models were predictive. This was also supported by their satisfactory 

Cite this paper
Hemal M.Soni,Popatbhai K.Patel,Mahesh T.Chhabria,Dharmraj N.Rana,Bhushan M.Mahajan,Pathik S.Brahmkshatriya, (2015) 2D-QSAR Study of a Series of Pyrazoline-Based Anti-Tubercular Agents Using Genetic Function Approximation. Computational Chemistry,03,45-53. doi: 10.4236/cc.2015.34006
References
- 1. Dye, C., Lonnroth, K., Jaramillo, E., Williams, B.G. and Raviglione, M. (2009) Trends in Tuberculosis Incidence and Their Determinants in 134 Countries. Bull, 87, 683-691.
http://dx.doi.org/10.2471/BLT.08.058453 - 2. Cole, S.T. and Riccardi, G. (2011) New Tuberculosis Drugs on the Horizon. Current Opinion in Microbiology, 14, 570-576.
http://dx.doi.org/10.1016/j.mib.2011.07.022 - 3. Udwadia, Z. F., Amale, R.A., Ajbani, K.K. and Rodrigues, C. (2012) Totally Drug-Resistant Tuberculosis in India. Clinical Infectious Diseases, 54, 579-581.
http://dx.doi.org/10.1093/cid/cir889 - 4. Chhabria, M.T., Mahajan, B.M. and Brahmkshatriya, P.S. (2011) QSAR Study of a Series of Acyl Coenzyme A (CoA): Cholesterol Acyltransferase Inhibitors Using Genetic Function Approximation. Medicinal Chemistry Research, 20, 1573-1580.
http://dx.doi.org/10.1007/s00044-010-9413-3 - 5. Buha, V.M., Rana, D.N., Chhabria, M.T., Chikhalia, K.H., Mahajan, B.M., Brahmkshatriya, P.S. and Shah, N.K. (2013) Synthesis, Biological Evaluation and QSAR Study of a Series of Substituted Quinazolines as Antimicrobial Agents. Medicinal Chemistry Research, 22, 4096-4109.
http://dx.doi.org/10.1007/s00044-012-0408-0 - 6. Tropsha, A. (2010) Best Practices for QSAR Model Development, Validation, and Exploitation. Molecular Informatics, 29, 476-488.
http://dx.doi.org/10.1002/minf.201000061 - 7. Ertan, T., Yildiz, I., Tekiner-Gulbas, B., Bolelli, K., Temiz-Arpaci, O., Ozkan, S., Yalcin, I. and Aki, E. (2009) Synthesis, Biological Evaluation and 2D-QSAR Analysis of Benzoxazoles as Antimicrobial Agents. European Journal of Medicinal Chemistry, 44, 501-510.
http://dx.doi.org/10.1016/j.ejmech.2008.04.001 - 8. Rana, D.N., Chhabria, M.T., Shah, N.K. and Brahmkshatriya, P.S. (2014) Pharmacophore Combination as a Useful Strategy to Discover New Antitubercular Agents. Medicinal Chemistry Research, 23, 370-381.
http://dx.doi.org/10.1007/s00044-013-0645-x - 9. Rana, D.N., Chhabria, M.T., Shah, N.K. and Brahmkshatriya, P.S. (2014) Discovery of New Antitubercular Agents by Combining Pyrazoline and Benzoxazole Pharmacophores: Design, Synthesis and Insights into the Binding Interactions. Medicinal Chemistry Research, 23, 2218-2228.
http://dx.doi.org/10.1007/s00044-013-0815-x - 10. Rogers, D. and Hopfinger, A.J. (1994) Application of Genetic Function Approximation to Quantitative Structure-Activity Relationships and Quantitative Structure-Property Relationships. Journal of Chemical Information and Computer Sciences, 34, 854-866.
http://dx.doi.org/10.1021/ci00020a020 - 11. Jaiswal, M., Khadikar, P.V., Scozzafava, A. and Supuran, C.T. (2004) Carbonic Anhydrase Inhibitors: The First QSAR Study on Inhibition of Tumor-Associated Isoenzyme IX with Aromatic and Heterocyclic Sulfonamides. Bioorganic & Medicinal Chemistry Letters, 14, 3283-3290.
http://dx.doi.org/10.1016/j.bmcl.2004.03.099 - 12. Roy, P.P. and Roy, K. (2007) On Some Aspects of Variable Selection for Partial Least Squares Regression Models. QSAR & Combinatorial Science, 27, 302-313.
- 13. Stanton, D.T. and Jurs, P.C. (1990) Development and Use of Charged Partial Surface Area Structural Descriptors in Computer Assissted Quantitative Structure Property Relationship Studies. Analytical Chemistry, 62, 2323-2329.
http://dx.doi.org/10.1021/ac00220a013
NOTES
*Corresponding author.





