Stem Cell Discovery
Vol.07 No.01(2017), Article ID:77869,26 pages
10.4236/scd.2017.71001
Mesenchymal Stromal Cells Derived from Human Embryonic Stem Cells, Fetal Limb and Bone Marrow Share a Common Phenotype but Are Transcriptionally and Biologically Different
Candida Vaz1, Betty Tan Bee Tee1, Delicia Yong2, Qian Yi Lee1, Vivek Tanavde1,3*
1Bioinformatics Institute, A*STAR (Agency for Science Technology and Research), Singapore
2Department of Medical Microbiology, Faculty of Medicine, University of Malaya, Kuala Lumpur, Malaysia
3Institute of Medical Biology, A*STAR (Agency for Science Technology and Research), Singapore

Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: January 1, 2017; Accepted: January 20, 2017; Published: January 30, 2017
ABSTRACT
Mesenchymal stromal cells (MSCs) can be obtained from several sources and the significant differences in their properties make it crucial to investigate the differentiation potential of MSCs from different sources to determine the optimal source of MSCs. We investigated if this biological heterogeneity in MSCs from different sources results in different mechanisms for their differentiation. In this study, we compared the gene expression patterns of phenotypically defined MSCs derived from three ontogenically different sources: Embryonic stem cells (hES-MSCs), Fetal limb (Flb-MSCs) and Bone Marrow (BM-MSCs). Differentially expressed genes between differentiated cells and undifferentiated controls were compared across the three MSC sources. We found minimal overlap (5% - 16%) in differentially expressed gene sets among the three sources. Flb-MSCs were similar to BM-MSCs based on differential gene expression patterns. Pathway analysis of the differentially expressed genes using Ingenuity Pathway Analysis (IPA) revealed a large variation in the canonical pathways leading to MSC differentiation. The similar canonical pathways among the three sources were lineage specific. The Flb-MSCs showed maximum overlap of canonical pathways with the BM-MSCs, indicating that the Flb-MSCs are an intermediate source between the less specialised hES-MSC source and the more specialised BM-MSC source. The source specific pathways prove that MSCs from the three ontogenically different sources use different biological pathways to obtain similar differentiation outcomes. Thus our study advocates the understanding of biological pathways to obtain optimal sources of MSCs for various clinical applications.
Keywords:
Mesenchymal Stromal Cells (MSCs), Human Embryonic Stem Cells Derived MSCs (hES-MSCs), Fetal Limb Derived MSCs (Flb-MSCs), Bone Marrow Derived MSCs (BM-MSCs), Ontogenically Different Sources, Source Specific Canonical Pathways

1. Introduction
Mesenchymal stromal cells (MSCs) are multipotent cells that were originally identified in bone marrow stroma [1] [2] [3] [4] . These were first described as mesenchymal stromal cells owing to their ability to differentiate into mesenchymal cells such as bone and cartilage. The minimum criteria for defining Mesenchymal stromal cells as defined by the International Society for Cellular Therapy (ISCT) include: plastic-adherence under standard culture conditions, presence of CD105, CD73, CD90 and absence of hematopoietic cell surface markers CD34, CD45, CD11a, CD19, and HLA-DR and most importantly the ability to differentiate into osteocytes, adipocytes, and chondrocytes in vitro [5] .
Mesenchymal stromal cells are known to differentiate into a variety of cell types including osteoblasts (bone cells), chondrocytes (cartilage cells), adipocytes (fat cells), myocytes (skeletal and cardiac muscle cells), and neurons (neural cells) [6] [7] .
MSCs are obtained from several adult mesenchymal tissue sources such as: bone marrow [8] , adipose tissue [9] , synovial membrane [10] , periosteum [11] and dental pulp [12] as well as from non-adult tissue sources such as the fetal tissue [13] , amniotic fluid [14] umbilical cord tissue (Wharton’s jelly) [15] and umbilical cord blood [16] .
Although MSCs can be obtained from different sources, they differ in their proliferative capacity and multilineage differentiation potential [17] . The proliferative capacity depends on factors such as the tissue source, age of the donor and culture conditions [18] [19] . Moreover, MSCs from different sources differentiate into a particular lineage more, or less efficiently depending on their source of origin. The bone marrow derived MSCs have better osteogenic differentiation potential as compared to the adipose derived MSCs that have better adipogenic differentiation potential [20] . The numbers of MSCs that can be isolated from different sources also vary enormously [21] .
Several biological studies have also shown superior biological properties of MSCs derived from neo-natal tissues over those derived from the adult tissues [22] . Another issue is that the MSCs isolated from various sources appear to be morphologically similar and display similar surface markers, yet they tend to have significant differences that cannot be attributed to the experimental variations.
Owing to these significant differences in the properties of the MSCs from different sources, it is crucial to investigate the differentiation potential of MSCs from different sources that is achieved through comparative analysis of their morphological and biological properties. Such comparative analysis of MSCs from different sources will help explain the transcriptional basis of heterogeneity observed in MSC from different sources and thus lead to more optimal selection of MSC from the right source for achieving a specific clinical outcome.
Some studies involve the comparison and analysis of properties such as the yield and differentiation potential, surface antigen expression, proliferation capacity, tolerance for aging and paracrine activity [20] [23] [24] . Such studies reveal the superiority and biological advantage of one source over the other and varied differentiation potential to different lineages.
There are a few studies involving comparisons of the transcriptome profiles of MSCs from different sources to identify the similarities and differences at the genetic level during differentiation into three major mesenchymal lineages [25] [26] [27] . Such studies provide a rational basis for assessing the qualities of MSCs from various sources.
However, till date there has been no comparative study of transcriptomes of ontogenically different sources and the mechanisms and pathways they follow for differentiation into similar lineages.
In our study, we compared the gene expression profiles of MSCs from three ontogenically different sources: Human Embryonic Stem cells (hES), Fetal limb (Flb) and Bone marrow (BM) at two time points of differentiation: Day 7 and Day 14/21. Currently there are no known markers that can identify MSCs at discrete stages of differentiation, therefore the only way to compare MSCs is at similar stage of differentiation. Even using cells from the same passage number does not guarantee that they will be at the same stage of differentiation. Therefore in this study we have used extent of differentiation as a method to compare these different MSC populations. In all 3 MSC populations, Day 7 was when differentiation was first detectable using histochemical stains. For Flb and BM MSCs differentiation was complete by day 14, whereas hES MSCs continued differentiation especially in the osteogenic lineage until Day 21. This was the basis of choosing the time points for comparison. The purpose of our study was to identify the similarities and differences at the genetic level as well as to explore the signaling pathways used by the MSCs from ontogenically different sources to differentiate into the similar mesodermal lineages. Since biological pathways rather than individual genes determine the phenotypic changes observed in cells, a deep understanding of the canonical pathways is essential. It is imperative to understand what pathways are used by the different sources to obtain similar differentiation products. Since MSCs have several clinical applications for treatment of various conditions such as: bone and cartilage diseases, cardiovascular diseases, liver diseases, autoimmune diseases and cancer [28] ; studies of this kind will enhance its therapeutic efficacy. Such studies will enable the application of MSCs as building blocks for molecular medicine [29] , in gene therapy [30] , and in regenerative medicine [31] .
2. Materials and Methods
2.1. Cell Culture
Human embryonic stem cell derived MSCs (hES-MSCs) were a gift from Choo A and Lim SK. Derivation of these cell lines have been described in a previous publication [32] . Cells were received at passage 11 and expanded for 2 passages in MSCGM BulletKit (Lonza, MD, USA).
Human fetal limb derived MSCs (Flb-MSCs) was a gift from Lim SK, who derived and characterized the cells [33] . The fetal limb derived MSC cell line was the F3lb cell line. Cells were received at passage 10 and expanded for 2 passages in MSCGM BulletKit (Lonza, MD, USA), with medium change every 3 - 4 days, and passage during 90% confluency (4 - 5 days) using TryPLE (Invitrogen, CA, USA) before differentiation.
Human bone marrow derived MSCs (BM-MSCs) were purchased frozen from Lonza (Gaithersburg, MD) at passage 2 [34] . These cells were thawed, cultured for an additional passage (passage 3), and pooled before being used in the experiments. All the MSCs confirmed to the ISCT definition of MSCs [5] .
hES-MSCs and Flb-MSCs were seeded at 5000 cells per cm2 in polystyrene cell culture flask (NUNC, NY, USA) and grown in 5% CO2, 37˚C incubator.
BM-MSCs were seeded at 3000 cells per cm2 and expanded on tissue culture treated?plastic dishes (Corning Life Sciences, Acton, MA). The MSC culture medium comprised of Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) or MesenPRO medium, 1xGlutaMAX, and 0.5 µg/mL gentamycin (all from Invitrogen). Cells were fed every alternate day and passaged using TryPLE, a recombinant protease (Invitrogen) when 80% confluence was reached. These samples were grown less than a month in culture (passage 3). For further details see Ng et al. 2008 paper [34] .
2.2. MSC Differentiation
After expansion, both hES-MSCs, Flb-MSCs were seeded in T75 culture flasks for RNA extraction and in 12-well plates for staining during differentiation of adipocytes, and osteocytes, and in pellet form in 15 ml polypropylene tubes for chondrocytes differentiation. Adipocytes and osteocytes grown in 12-well plates were used for Oil Red O and Von Kossa Staining respectively, while Alcian blue staining was done for chondrocytes pellet grown in polypropylene tube.
MSCs were seeded at 1.8 × 104 cells per cm2 for adipogenic differentiation, 0.27 × 104 cells per cm2 for osteogenic differentiation, and in pellet size of 50 × 104 cells for chondrogenic differentiation. Trilineage differentiation was done using hMSC adipogenic, osteogenic, and chondrogenic differentiation BulletKits with recombinant human TGF-β3 (Lonza, MD, USA), according to manufacturer’s protocol. Media was changed every 2 - 4 days.
BM-MSCs were expanded in culture and seeded in T125 flasks for differentiation to either adipocytes, osteocytes or chondrocytes at different passages. Cells were also seeded in parallel on chamber slides for differentiation into osteogenic and adipogenic lineages and were stained with Von Kossa and Oil red O, respectively. MSCs were seeded at a density of 20,000 cells per cm2 in the presence of adipogeneic differentiation media containing 0.5 mM isobutyl-methylxanthine (Aldrich, Milwaukee, WI), 10 µM bovine insulin, 1 µM dexamethasone, and 200 µM indomethacin (all from Sigma-Aldrich, St Louis, MO). Osteogeneic differentiation (seeding density 5000 cells per cm2) media comprised of MSC growth media with 10 mM glycerol-2-phosphate, 50 µM L-ascorbic acid, and 100 nM dexamethasone (all from Sigma-Aldrich).
Chondrogenic differentiation of MSCs was induced through aggregate culture. Aliquots of 2.5 × 105 cells were centrifuged at 160 g for 5 minutes in 15 mL polypropylene conical tubes to form pellets, which were cultured in medium containing high glucose DMEM supplemented with 4 mM proline, 50 µg/mL ascorbic acid, 1% ITS-Premix (BD Biosciences, San Jose, CA), 1 mM sodium pyruvate, and 0.1 µM dexamethasone for 15 days in the presence of 10 ng/mL of TGF-β3 (R&D Systems, Minneapolis, MN). Media was changed every 3 days. For further details see Ng et al. 2008 paper [34] .
2.3. RNA Extraction
Total RNA of the undifferentiated MSC, adipocytes, and osteocytes of hES-MSCs and Flb-MSCs was harvested using 1 ml of Trizol (Invitrogen, CA, USA)/10 cm2 area. Chondrocyte pellets were treated with 0.25% of collagenase type II in DMEM high glucose medium (Invitrogen, CA, USA) for 1 - 2 hr at 37˚C, centrifuged at 1200 g for 5 min, and the supernatant replaced with 300 ul of Trizol per pellet. All samples were stored in Trizol at −80˚C until RNA extraction.
RNA extraction from samples in Trizol was done using chloroform and purified with RNeasy kit (Qiagen, Netherlands). Concentration of RNA was determined using Nanodrop (Agilent Technologies, CA, USA) and the RNA integrity measured using RNA Nano LabChip on Agilent Bioanalyzer (Agilent Technologies, CA, USA). Only high quality total RNA samples of OD260/OD280 > 1.8 and RNA Integrity Number (RIN) of > 8, were used for microarray.
BM-MSC Cells (5 × 106) from undifferentiated MSCs as well as their differentiated progeny were dissolved in Trizol and stored at −80˚C for RNA extraction. RNA extraction from Trizol samples was carried out using the RNA easy kit (Qiagen, Valencia, CA).
2.4. Gene Expression Microarray
For the hES-MSCs and Flb-MSCs, RNA amplification was done using Illumina TotalPrepTM RNA amplification kit (Ambion, TX, USA), according to manufacturer’s instructions. 750 ng of biotin-labelled cRNA for hybridization was randomly loaded on to Illumina Sentrix BeadChip Array human Ref-8v3 bead chips (Illumina, San Diego, CA).
After 16 hr of hybridization, the Beadchips were washed, Cy3-labeled, and scanned with an Illumina Bead array reader confocal scanner and the data uploaded to GenomeStudio (Illumina, CA, USA) for background subtraction and conversion into Partek file for data analysis on Partek Genomics Suite.
For BM-MSC, RNA amplification was performed using 500 ng of total RNA with the Illumina TotalPrepTM RNA amplification kit (Ambion, TX, USA) following the manufacturer’s instructions. The biotin-labeled RNA was hybridized to a human Ref-8v1 bead chip (Illumina, San Diego, CA). Hybridized arrays were scanned with an Illumina Bead array reader confocal scanner and data uploaded to GenomeStudio (Illumina, CA, USA) for background subtraction and conversion into Partek file for data analysis on Partek Genomics Suite.
2.5. Details of the Microarray Datasets Used for the Analysis
The microarray datasets comprised of the three ontogenically different sources (hES-MSCs, Flb-MSCs and BM-MSCs) differentiated into the three lineages (Adipocytes (A), Chondrocytes (C) and Osteocytes (O)) at Day 7 and Day 14/21 as well as their undifferentiated form that was set as the control (Basal (B0) at Day 0). For each source there were at least two biological replicates and for each biological replicate there were technical replicates. The total numbers of replicates were: 6 - 14 for the hES-MSCs, 4 - 7 for the Flb-MSCs and 6 for BM-MSCs.
Data availability: The microarray data of hES-MSCs and Flb-MSCs have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO Series accession number GSE100752. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100752
GSE100752 comprises of two SubSeries: GSE100748: microarray data of hES- MSCs; GSE100749: microarray data of Flb-MSCs.
The microarray data of BM-MSCs is deposited in ArrayExpress and is accessible through the accession number E-TABM-318 https://www.ebi.ac.uk/arrayexpress/experiments/E-TABM-318/.
2.6. Normalisation, Quality Control and Obtaining Differentially Expressed Genes
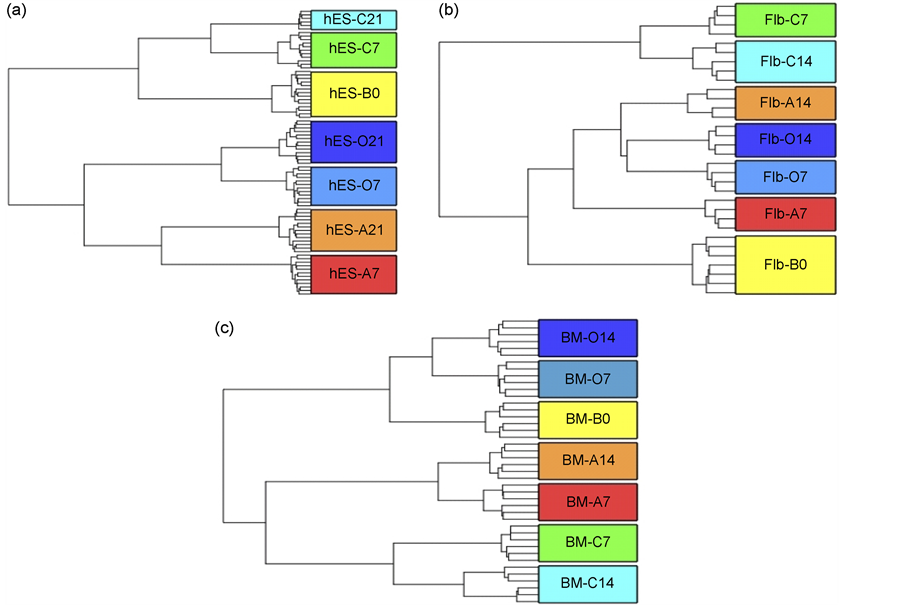
The microarray data was imported in Partek, and normalized using “quantile” normalization. The batch effect due to different chips (sentrix barcode), also known as random effect, was removed and quality control check was carried out to make sure the replicates clustered according to the lineages they were derived from. The hierarchical clustering plots were generated for each of the three sources to make sure the clustering was optimal for further processing (Figure 1).
To obtain the differentially expressed genes between the differentiated sample and the undifferentiated basal (B0) control for each of the three lineages (Adipocytes, Chondrocytes and Osteocytes) and for each of the sources (hES, Flb and BM) at Day 7 and Day 14/21, a multiple factor Analysis of Variance (ANOVA)
Figure 1. Hierarchical clustering plots showing the clustering of the Basal (undifferentiated) (B0) and the three differentiated lineages: Adipocytes (A), Chondrocytes (C) and Osteocytes (O) sample replicates at Day 7 (7) and Day 14/21 (14/21). (a) Human Embryonic Stem Cells (hES) derived MSCs plot (b) Fetal Limb (Flb) derived MSCs plot (c) Bone Marrow derived MSCs (BM) plot. The plot shows that MSCs at different stages of differentiation cluster based on their stage of differentiation. The replicates cluster together in this plot as expected indicating similarity in their transcriptomes.
was performed. The type of lineage, day of differentiation, interaction of type of lineage and day of differentiation as well as the sentrix barcode (random effect) were taken as ANOVA factors. A significant gene list was obtained using a p-value with False Discovery Rate (FDR) < 0.05 cut-off and a fold-change cut-off of 2. The details of the DE genes obtained are given in the supplementary files (Supplementary Files 1 - 3).
2.7. Obtaining HUGO Gene Nomenclature Committee (HGNC) Approved Symbols
Owing to the difference in the versions of the Illumina platforms, the symbols of the differentially expressed genes obtained were run through the “Multi-symbol checker” tool to check if they were HGNC approved symbols. In case if a symbol was not the current approved symbol but a previously used symbol, it was replaced by the currently approved symbol.
2.8. Canonical Pathways Analysis
The HGNC approved differentially expressed gene lists obtained were uploaded into Ingenuity pathway analysis software (IPA). A core analysis for each of the lists in context of canonical pathways was carried out in IPA. IPA reports the enriched pathways for the dataset and assigns a ratio and a significance score derived from the p-value, using the right-tailed Fisher exact test. The ratio is calculated by taking the number of genes from the user’s dataset that participate in a canonical pathway and dividing it by the total number of genes in that canonical pathway. The p-value measures how likely the observed association between a specific pathway and the dataset would be if it was only due to random chance. The smaller is the p-value, the more is the confidence of the corresponding pathway being associated with the user’s dataset. Pathways having higher ratios and lower p-values are more likely to explain the phenotype of the dataset than others. Significance score is the negative log of the p-value. A threshold score of p-value 0.01 corresponding to a significance score of 2 was used.
The significantly enriched pathways were compared for each lineage (differentiated cell type) at the two stages (Day 7 and Day 14/21) among the three sources to identify the ones commonly significant (negative log p-value = 2) among all the three sources or any of the two sources and also specific to each source.
3. Results
3.1. Microarray Datasets and Obtaining Differentially Expressed Genes
The Microarray datasets comprised of the three ontogenically different sources (hES-MSCs, Flb-MSCs and BM-MSCs) differentiated into the three lineages (Adipocytes (A), Chondrocytes (C) and Osteocytes (O)) at Day 7 and Day 14/21 as well as their undifferentiated form that was set as the control (Basal (B0) at Day 0).
The microarray data was imported in Partek, and normalized using “quantile” normalization. The hierarchical clustering plots were generated for each of the three sources to make sure the clustering was optimal for further processing (Figure 1).
To obtain the differentially expressed genes between the differentiated sample and the undifferentiated basal (B0) control for each of the three lineages (Adipocytes, Chondrocytes and Osteocytes) and for each of the sources (hES, Flb and BM) at Day 7 and Day 14/21, a multiple factor Analysis of Variance (ANOVA) was performed. A significant gene list was obtained using a p-value with False Discovery Rate (FDR) < 0.05 cut-off and a fold-change cut-off of 2. For detailed information see the “Materials and Methods” section. The details of the DE genes obtained are given in the supplementary files (Supplementary Files 1 - 3).
3.2. MSCs from the Three Ontogenically Different Sources, Differ in Their Growth Kinetics and Differentiation Abilities
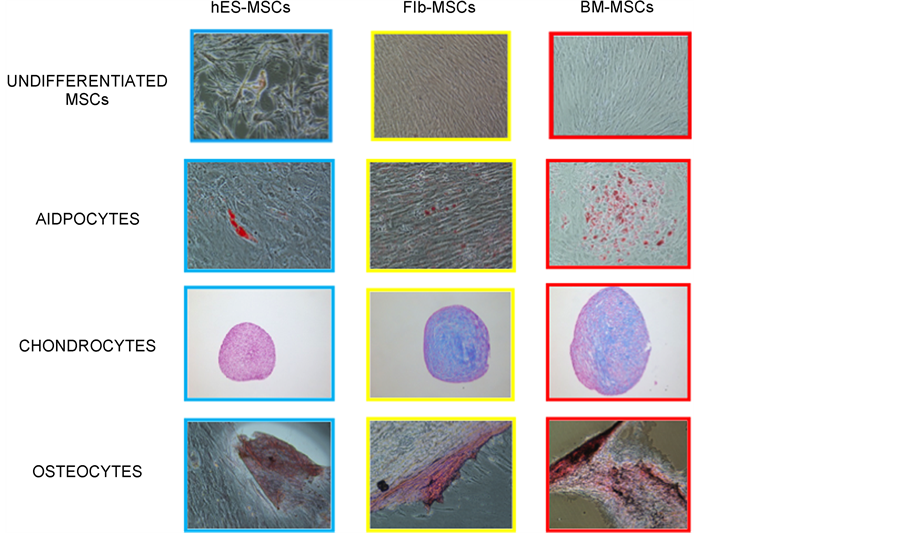
The hES-MSCs take the maximum time to differentiate (around 21 days), as compared to the Flb-MSCs and BM-MSCs (around 14 days). However, the hES-MSCs and Flb-MSCs can be cultured upto 18 to 19 passages as compared to the BM-MSCs that can be cultured only up to 6 - 7 passages. The doubling time for Flb-MSCs is the least (2 - 3 days) as compared to the hES-MSCs (3 - 4 days) and the BM-MSCs (4 - 5 days) Table 1. The hES-MSCs show poorer differentiation ability into the three lineages (Adipocytes, Chondrocytes and Osteocytes) as compared to the Flb-MSCs and BM-MSCs (Figure 2).
Table 1. MSCs from the three ontogenically different sources differ in their growth kinetics.
Figure 2. MSCs from three ontogenically different sources, differ in their differentiation abilities. The figure shows that the three sources differ in their differentiation ability. The Human Embryonic Stem Cells (hES) derived MSCs have the poorest trilineage differentiation potential.
3.3. Minimal Overlap Found on Comparing the Differentially Expressed Genes among the Three Ontogenically Different Sources
We found that the differentially expressed genes were highly dissimilar between the cells differentiated from hES-MSCs, Flb-MSCs and BM-MSCs.
In adipocytes at Day 7, there were 708 differentially expressed genes from hES-MSCs, 1155 genes from Flb-MSCs and 769 genes from BM-MSCs. Of these only 106 genes were similar among all three sources of MSCs (Figure 3a(i)). In adipocytes at Day 14/21, there were 801 differentially expressed genes from hES-MSCs, 678 genes from Flb-MSCs and 920 genes from BM-MSCs. Of these, only 68 genes were similar among all three sources of MSCs (Figure 3a(ii)).
In chondrocytes at Day 7, there were 709 differentially expressed genes from hES-MSCs, 1305 genes from Flb-MSCs and 887 genes from BM-MSCs. Of these, only 113 genes were similar among all three sources of MSCs (Figure 3b(i)).
In chondrocytes at Day 14/21, there were 1179 differentially expressed genes from hES-MSCs, 1147 genes from Flb-MSCs and 1613 genes from BM-MSCs. Of these, only 86 genes were similar among all three sources of MSCs (Figure 3b(ii)).
Figure 3. Comparison of the differentially expressed genes among the three sources. (a) for Adipocyte differentiation, (b) for Chondrocyte differentiation, (c) for Osteocyte differentiation. (i) at Day 7 (ii) at Day 14/21. The figure shows very less extent of overlap among the differentially expressed genes of the three sources and for the three differentiated lineages.
Finally in osteocytes at Day 7, there were 655 differentially expressed genes from hES-MSCs, 786 genes from Flb-MSCs and 473 genes from BM-MSCs. Of these, only 68 genes were similar among all three sources of MSCs (Figure 3c(i)).
In osteocytes at Day 14/21, there were 904 differentially expressed genes from hES-MSC, 885 genes from Flb-MSC and 602 genes from BM-MSCs. Of these, only 93 genes were similar among all three sources of MSCs (Figure 3c(ii)).
3.4. Known Lineage Specific Markers
We looked at expression of known lineage specific markers in the differentially expressed gene lists to check to what extent they were regulated across the three sources. Tables 2-4 comprise of the fold change values of the lineage specific markers across the three sources.
Table 2. Fold change in expression of the known lineage specific markers across the three sources for adipogenesis.
p-value with False Discovery Rate (FDR) < 0.05.
3.4.1. Adipogenesis Markers
Out of the 25 known adipogenesis markes, only one was found to be significantly regulated in hES-MSC source at both Day 7 and Day 21. The Flb-MSC source had 8 adipogenesis markers at both Day 7 and Day 14. The BM-MSC source had 6 and 9 adipogenesis markers at Day 7 and Day 14 respectively (Table 2). This indicates that the hES-MSC source differentiates poorly into adipocytes as compared to the Flb-MSC and the BM-MSC sources.
3.4.2. Chondrogenesis Markers
Out of the 28 known chondrogenesis markers, The hES-MSC source had 4 and
Table 3. Fold change in expression of the known lineage specific markers across the three sources for chondrogenesis.
p-value with False Discovery Rate (FDR) < 0.05.
2 chondrogenesis markers at Day 7 and Day 21 respectively. The Flb-MSC source had 8 and 10 chondrogenesis markers at Day 7 and Day 14 respectively. The BM-MSC source had 9 and 11 chondrogenesis markers at Day 7 and Day 14 respectively (Table 3). This indicates that the hES-MSC source differentiates poorly into chondrocytes as compared to the Flb-MSC and the BM-MSC sources.
3.4.3. Osteogenesis Markers
Out of the 26 known osteogenesis markers, The hES-MSC source had 4 and 5 osteogenesis markers at Day 7 and Day 21 respectively. The Flb-MSC source had 6 and 5 osteogenesis markers at Day 7 and Day 14 respectively. The BM-MSC source had 4 and 5 osteogenesis markers at Day 7 and Day 14 respectively (Table 4).
Table 4. Fold change in expression of the known lineage specific markers across the three sources for osteogenesis.
p-value with False Discovery Rate (FDR) < 0.05.
3.5. The Differentially Expressed Genes Are Manifested into Different Canonical Pathways
The HGNC approved differentially expressed gene lists obtained were uploaded into Ingenuity pathway analysis software (IPA). A core analysis for each of the lists in context of canonical pathways was carried out in IPA. IPA reports the enriched canonical pathways for the dataset and assigns a ratio and a significance score derived from the p-value, using the right-tailed Fisher exact test (For details see the “Materials and Methods” section).
The gene expression data showed that trilineage differentiation from hES-MSC, Flb-MSC and BM-MSC yield certain differences and similarity to the differentially regulated genes, causing changes in the signaling pathways regulated. Since all the three MSC sources ultimately differentiate into the three lineages, we expected some canonical pathways to be similar amongst them and some to be very specific. For this purpose the comparison among the enriched canonical pathways was carried out for each lineage (differentiated cell type) at the two different stages (Day 7 and Day 14/21) among the three sources to identify the ones commonly significant (negative log p-value = 2) among all the three sources or any of the two sources and also specific to each source.
3.5.1. Adipogenesis Day 7
The canonical pathways significant in common between MSCs derived from all three sources were 13. Among these 13, the most significant ones were “Hepatic Fibrosis/Hepatic Stellate Cell Activation”, “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis”, “LXR/RXR Activation”, “Granulocyte Adhesion and Diapedesis”, “HMGB1 Signaling”, “Acute Phase Response Signaling” and “Adipogenesis pathway”.
Only 6 canonical pathways were significant in common for hES and Flb whereas 11 were significant in common between hES and BM, indicating that the canonical pathways of the hES MSC source are different from those of the Flb and BM MSC sources for adipogenesis at day 7. Among the common significant pathways the “IL-17A Signaling in Fibroblasts”, “PPAR Signaling” and “IL-6 Signaling” were the most significant pathways for hES and Flb only and the “Mitotic Roles of Polo-Like Kinase”, “Aryl Hydrocarbon Receptor Signaling”, “Colorectal Cancer Metastasis Signaling” and “Growth Hormone Signaling” were the most significant pathway for hES and BM only.
The canonical pathways significant in common between MSCs derived from Flb and BM only were 12. Among these 12, the most significant ones were “LPS/ IL-1 Mediated Inhibition of RXR Function” and “Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis”.
The canonical pathways significant in hES only were 29; among these the most significant pathways were “Interferon Signaling”, “IL-10 Signaling” and “Toll-like Receptor Signaling”.
The canonical pathways significant in Flb only were 6; among these the most significant pathway was the “Epithelial Adherens Junction Signaling”.
The canonical pathways significant in BM only were 31; among these the most significant pathway were “Cell Cycle: G2/M DNA Damage Checkpoint Regulation”, “Actin Nucleation by ARP-WASP Complex” and “Clathrin-mediated Endocytosis Signaling” (Supplementary File 4 Sheet 1).
3.5.2. Adipogenesis Day 14/21
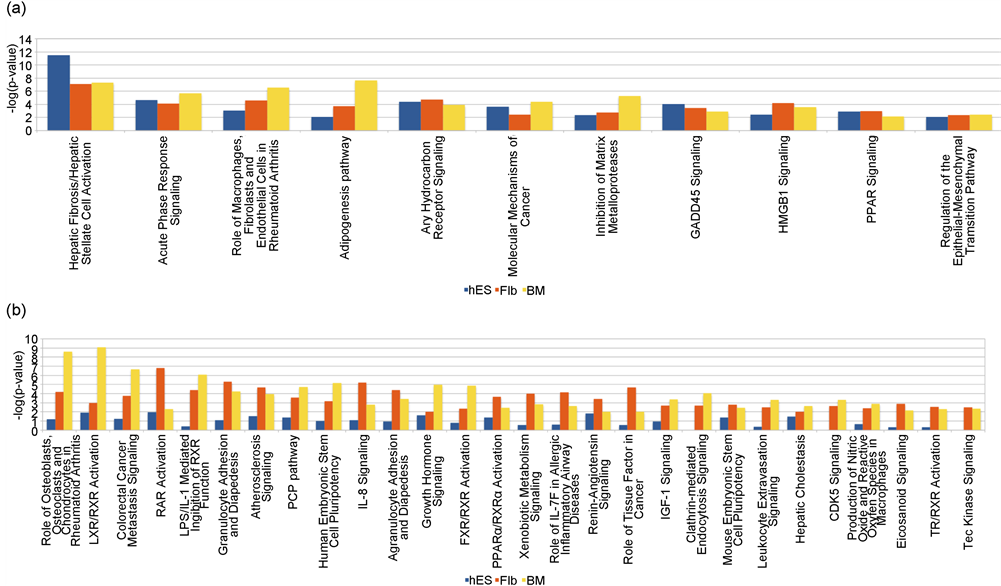
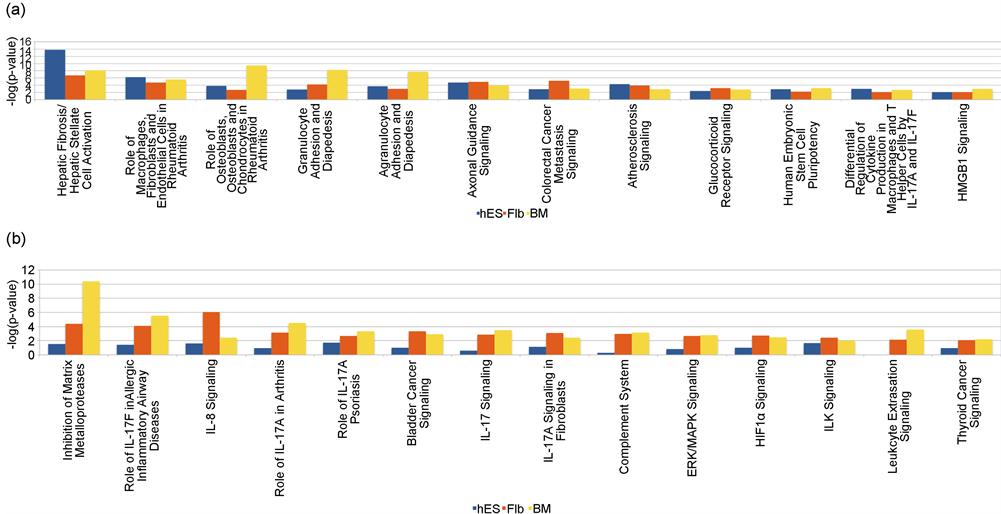
The canonical pathways significant in common between MSCs derived from all three sources were 11 (Figure 4(a)). The canonical pathways significant in common between MSCs derived from hES and Flb only and hES and BM only were 9 and 5 respectively, indicating that the canonical pathways of the hES MSC source are different from those of the Flb and BM MSC sources for adipogenesis at day 14/21. Among the commonly significant pathways the “IL-6 Signaling”, “Glucocorticoid Receptor Signaling”, “Pancreatic Adenocarcinoma Signaling” and “IL-17A Signaling in Fibroblasts” were the most significant pathways in common between MSCs derived from hES and Flb only and the “Mitotic Roles of Polo-Like Kinase”, “Axonal Guidance Signaling” and “Cyclins and Cell Cycle Regulation” were the most significant pathways in common between MSCs derived from hES and BM only.
Figure 4. Comparison of the canonical pathways among the three sources for adipogenesis at Day 14/21. (a) Significant for all the three sources: The canonical pathways significant in common between MSCs derived from all three sources were 11. Among these 11, the most significant ones were “Hepatic Fibrosis/Hepatic Stellate Cell Activation”, “Acute Phase Response Signaling”, “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis”, “Adipogenesis pathway” and “Aryl Hydrocarbon Receptor Signaling” (b) Significant for Flb and BM MSCs only: The canonical pathways significant in common between MSCs derived from Flb and BM only were 28. Among these 28, the most significant ones were “Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis”, “LXR/RXR Activation”, “Colorectal Cancer Metastasis Signaling and “RAR Activation”.
The canonical pathways significant in common between MSCs derived from Flb and BM only were 28 (Figure 4(b)).
The canonical pathways significant in hES derived MSCs only were 19; among these the most significant pathways were “Cell Cycle Control of Chromosomal Replication”, “Estrogen-mediated S-phase Entry”, “Cell Cycle: G1/S Checkpoint Regulation” and “IL-10 Signaling”.
The canonical pathways significant in Flb derived MSCs only were 30; among these the most significant pathway were “IL-17 Signaling”, “NRF2-mediated Oxidative Stress Response” and “Role of IL-17A in Arthritis”.
The canonical pathways significant in BM derived MSCs only were 28; among these the most significant pathway were “Glioblastoma Multiforme Signaling”, “Dopamine Receptor Signaling”, and “Wnt/β-catenin Signaling” (Supplementary File 4 Sheet 2).
3.5.3. Chondrogenesis Day 7
The canonical pathways significant in common between MSCs derived from all three sources were 20. Among these 20, the most significant ones were “Hepatic Fibrosis/Hepatic Stellate Cell Activation”, “Colorectal Cancer Metastasis Signaling”, “Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis”, “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis” and “Inhibition of Matrix Metalloproteases”.
The canonical pathways commonly significant for hES and Flb only and hES and BM only were 21 and 17 respectively. Among the commonly significant pathways the “Granulocyte/Agranulocyte Adhesion and Diapedesis” and “Differential Regulation of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F” were the most significant pathways between hES and Flb and “Role of Tissue Factor in Cancer”, “Mitotic Roles of Polo-Like Kinase”, “Molecular Mechanisms of Cancer”, “Ovarian Cancer Signaling” and “Cyclins and Cell Cycle Regulation”, were the most significant pathways between hES and BM respectively.
The canonical pathways significant in common between MSCs derived from Flb and BM only were 9. Among these 9, the most significant ones were “Thyroid Cancer Signaling”, “Signaling by Rho Family GTPases”, “HMGB1 Signaling” and “IL-10 Signaling”.
The canonical pathways significant in hES only were 15; among these the most significant pathways were “Estrogen-mediated S-phase Entry” and “TNFR2 Signaling”.
The canonical pathways significant in Flb only were 23; among these the most significant pathway were “Graft-versus-Host Disease Signaling” and “Communication between Innate and Adaptive Immune Cells”.
The canonical pathways significant in BM only were 28; among these the most significant pathways were “Regulation of Cellular Mechanics by Calpain Protease” and “RhoA Signaling” (Supplementary File 4 Sheet 3).
3.5.4. Chondrogenesis Day 14/21
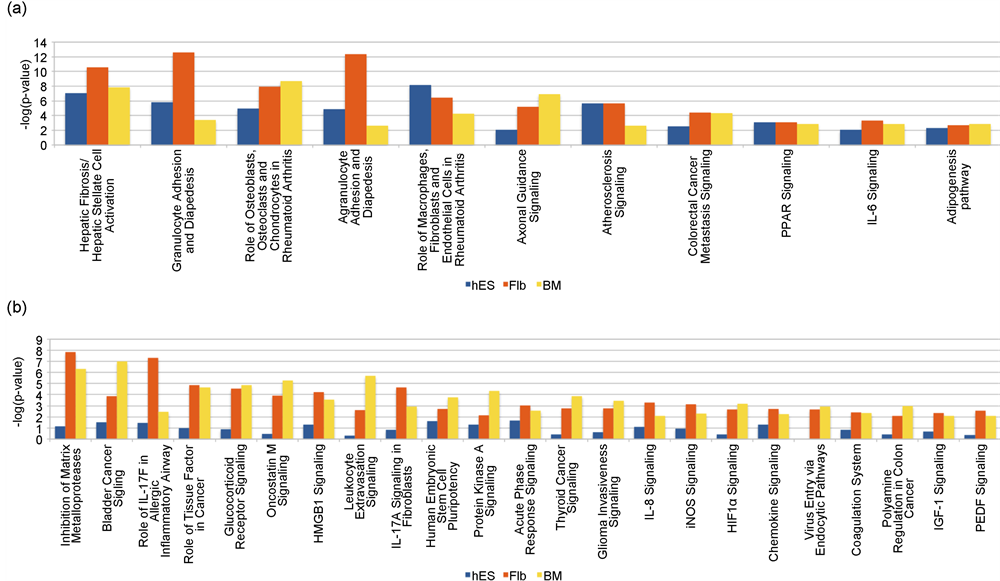
The canonical pathways significant in common between MSCs derived from all three sources were 11 (Figure 5(a)). The canonical pathways commonly significant for hES and Flb only and hES and BM only were 12 and 16 respectively. Among the commonly significant pathways the “LXR/RXR Activation”, “Differential Regulation of Cytokine Production in Macrophages and T Helper Cells by IL-17A and IL-17F” and “Differential Regulation of Cytokine Production in Intestinal Epithelial Cells by IL-17A and IL-17F” were the most significant pathways for hES and Flb only and the “Mitotic Roles of Polo-Like Kinase”, “Cell Cycle: G2/M DNA Damage Checkpoint Regulation”, “Aryl Hydrocarbon Receptor Signaling”, “Cyclins and Cell Cycle Regulation” and “Molecular Mechanisms of Cancer” were the most significant pathways for hES and BM only.
The canonical pathways significant in common between MSCs derived from Flb and BM only were 23 (Figure 5(b)).
There were 8 canonical pathways significant for hES derived MSCs only, among these the most significant ones were “Interferon Signaling” and “ATM Signaling”.
Figure 5. Comparison of the canonical pathways among the three sources for chondrogenesis at Day 14/21. (a) Significant for all the three sources: The canonical pathways significant in common between MSCs derived from all three sources were 11. Among these 11, the most significant ones were “Hepatic Fibrosis/Hepatic Stellate Cell Activation”, “Granulocyte Adhesion and Diapedesis”, “Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis”, “Agranulocyte Adhesion and Diapedesis” and “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis” (b) Significant for Flb and BM MSCs only: The canonical pathways significant in common between MSCs derived from Flb and BM only were 23. Among these 23, the most significant ones were “Inhibition of Matrix Metalloproteases”, “Bladder Cancer Signaling”, “Role of IL-17F in Allergic Inflammatory Airway Diseases”, “Role of Tissue Factor in Cancer” and “Glucocorticoid Receptor Signaling”.
23 canonical pathways were significant for Flb derived MSCs only; among these the most significant pathways were “Role of IL-17A in Arthritis” and “IL-17A Signaling in Gastric Cells”.
The canonical pathways significant for BM only were 58; among these the most significant pathways were “Regulation of Cellular Mechanics by Calpain Protease”, “Integrin Signaling”, “Actin Cytoskeleton Signaling” and “EIF2 Signaling” (Supplementary File 4 Sheet 4).
3.5.5. Osteogenesis Day 7
The canonical pathways significant in common between MSCs derived from all three sources were 15. Among these 15, the most significant ones were “Hepatic Fibrosis/Hepatic Stellate Cell Activation”, “Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis”, “Agranulocyte Adhesion and Diapedesis”, “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis”, “Granulocyte Adhesion and Diapedesis”, and “Inhibition of Matrix Metalloproteases”.
The canonical pathways commonly significant for hES and Flb only and hES and BM only were 8 and 3 respectively. Among the commonly significant pathways the “IL-8 Signaling”, “Colorectal Cancer Metastasis Signaling” and “Airway Pathology in Chronic Obstructive Pulmonary Disease” were the most significant in common between hES and Flb derived MSCs only and “Human Embryonic Stem Cell Pluripotency”, “LXR/RXR Activation” and “Regulation of the Epithelial-Mesenchymal Transition Pathway” were the most significant for hES and BM only.
The canonical pathways significant in common between MSCs derived from Flb and BM only were 5 namely, “Role of IL-17F in Allergic Inflammatory Airway Diseases”, “Leukocyte Extravasation Signaling”, “Role of Tissue Factor in Cancer”, “Complement System” and “Aldosterone Signaling in Epithelial Cells”.
The canonical pathways significant for hES only were 15; among these the most significant pathways was “Wnt/β-catenin Signaling” and “Epithelial Adherens Junction Signaling”.
The canonical pathways significant for Flb only were 20; among these the most significant pathways were “Cell Cycle: G2/M DNA Damage Checkpoint Regulation”, “GADD45 Signaling” and “Mitotic Roles of Polo-Like Kinase”.
The canonical pathways significant for BM only were 11; among these the most significant pathway was “ERK/MAPK Signaling” (Supplementary File 4 Sheet 5).
3.5.6. Osteogenesis Day 14/21
The canonical pathways significant in common between MSCs derived from all three sources were 12 (Figure 6(a)). The canonical pathways significant in common between MSCs derived from hES and Flb only and hES and BM only were 5 and 3 respectively. The common 5 significant pathways for hES and Flb only were the “Aryl Hydrocarbon Receptor Signaling”, “Adipogenesis pathway”,
Figure 6. Comparison of the canonical pathways among the three sources for osteogenesis at Day 14/21. (a) Significant for all the three sources: The canonical pathways significant in common between MSCs derived from all three sources were 12. Among these 12, the most significant ones were “Hepatic Fibrosis/Hepatic Stellate Cell Activation”, “Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis”, “Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis”, “Granulocyte Adhesion and Diapedesis” and “Agranulocyte Adhesion and Diapedesis” (b) Significant for Flb and BM MSCs only: The canonical pathways significant in common between MSCs derived from Flb and BM only were 14. Among these 14, the most significant were “Inhibition of Matrix Metalloproteases”, “Role of IL-17F in Allergic Inflammatory Airway Diseases” and “IL-8 Signaling”.
“Wnt/β-catenin Signaling”, “Ovarian Cancer Signaling” and “Regulation of the Epithelial-Mesenchymal Transition Pathway” and the common 3 significant pathways for hES and BM only were “IL-17A Signaling in Gastric Cells”, “Acute Phase Response Signaling” and “Molecular Mechanisms of Cancer”.
The canonical pathways significant in common between MSCs derived from Flb and BM only were 14 (Figure 6(b)).
The canonical pathways significant for hES only were 14; among these the most significant pathways was “Differential Regulation of Cytokine Production in Intestinal Epithelial Cells by IL-17A and IL-17F” and “iNOS Signaling”.
The canonical pathways significant for Flb only were 26; among these the most significant pathways were “Role of Tissue Factor in Cancer” and “Eicosanoid Signaling”.
The canonical pathways significant for BM only were 11; among these the most significant ones were “LXR/RXR Activation” and “NRF2-mediated Oxidative Stress Response” (Supplementary File 4 Sheet 6).
4. Discussion
MSCs can be obtained from several adult mesenchymal tissue sources as well as neonatal tissue sources. Though MSCs from different sources are morphologically similar, display similar surface markers and can eventually differentiate into the three mesenchymal lineages: Adipocytes, Chondrocytes and Osteocytes; they differ in their proliferative capacity and differentiation potential. The multitude of sources available creates the necessity of finding the most appropriate MSC source.
Several papers have reported the superiority of one source over the other for one particular lineage [20] [22] [23] [24] . A few studies have provided a rational basis of accessing the qualities of MSCs from various sources by comparing the transcriptome profile of MSCs from different sources and identifying their similarities and differences at the genetic level [25] [26] [27] . However, none of the studies so far has reported a comparison of MSC sources from different ontogeny using a systematic study to compare their transcriptomes.
In this study, we compared the gene expression profiles of MSCs from three ontogenically different sources: human embryonic stem cells, fetal limb and bone marrow. MSCs are also known for their ability to secrete paracrine factors that promote the recruitment of host cells. These paracrine factors are very important in tissue regeneration and repair of MSCs. Transcriptome profiling provides an effective way to measure global expression of these paracrine factors. Therefore in this study we have compared the transcriptomes of MSCs from different sources. The aim of our study was not only to look for the similarities and differences between the transcriptome of these MSCs, but also to explore all the pathways used by the MSCs to differentiate into the three lineages.
The choice of MSC sources depended on their availability, growth kinetics and differentiation potential. hES-MSCs have an unlimited supply, short doubling time (3 - 4 days) and can be passaged for longer periods (18 - 19 passages). BM-MSCs are a gold standard and are widely used owing to their excellent differentiation abilities. Flb-MSCs are an intermediate between the hES-MSCs and BM-MSCs; they have a short doubling time (2 - 3 days), can be passaged for longer periods (18 - 19 passages). Flb-MSCs are similar to BM-MSCs in terms of their growth and differentiation abilities, but senesce much later and can be passaged for 18 - 19 passages. Thus these would be ideal model system for studying MSCs and comparing their characteristics to BM-MSCs.
We first compared the gene expression profiles of MSCs from all 3 sources. Identification of differentially expressed genes with respect to the undifferentiated control was carried out at Day 7 and Day 14/21. The lists of the differentially expressed gene were compared across the three sources for the three lineages. We found that the differentially expressed genes were highly dissimilar with very less overlap (5% - 16%) among the three sources.
We compared the expression of known lineage specific markers compiled from literature to check to what extent were they regulated across the three sources. The presence of few known markers in the list of significant differentially expressed gene lists of adipocytes, chondrocytes and osteocytes indicates that the hES-MSC source differentiates poorly into the three lineages as compared to the Flb-MSC and the BM-MSC sources. The poor multilineage differentiation ability of hES-MSCs by current protocols was also observed in a study comparing bone marrow and embryonic stem cells derived MSCs. The authors observed that the embryonic stem cells derived MSCs were more proliferative and had a higher capability of immunomodulation, but were less inducible for differentiation as compared to the BM-MSCs [35] .
Since biological pathways and networks rather than individual genes determine the phenotypic changes observed in cells, we studied the canonical pathways these differentially expressed genes were involved in.
For this purpose, the differentially expressed gene lists were uploaded into Ingenuity pathway analysis tool. IPA reports the enriched pathways for the dataset and assigns a ratio and a significance score derived from the p-value. Owing to the difference in the differentially expressed genes, the canonical pathways also exhibited a large variation.
The similar canonical pathways among the three sources were lineage specific. For adipocytes the most significant common pathways were: “Acute Phase Response Signaling”, “Adipogenesis pathway”, and HMGB1 Signaling. For chondrocytes the most significant common pathways were: the “Axonal Guidance Signaling”, “Colorectal Cancer Metastasis Signaling”, Inhibition of Matrix Metalloproteases” and “Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis”. For osteocytes the common pathways were: “Agranulocyte Adhesion and Diapedesis”, “Granulocyte Adhesion and Diapedesis”, “Inhibition of Matrix Mettaloproteases” and Role of Osteoblasts, Osteoclasts and Chondrocytes in Rheumatoid Arthritis. The Flb-MSCs showed maximum overlap of canonical pathways with the BM-MSCs, indicating that the Flb-MSCs is an intermediate source between the less specialised hES-MSC source and the more specialised BM-MSC source.
This source specific utilization of canonical pathways for differentiation suggests that the MSCs from the three ontogenically different sources use different pathways and networks to obtain similar differentiation products.
These observations explain the heterogeneity of MSCs observed at the cellular level arise from differences in the transcriptome of MSC from different sources. This heterogeneity is important in choosing MSC from the right source for a specific therapeutic application. This is especially relevant since certain MSC like dental pulp MSC can only be harvested at limited points and need to be banked. To improve the utilization of MSCs for cellular therapy it is also essential to have a standard/reference material to overcome the inconsistencies of clinical trials that arise due to several variable factors. The minimal criteria recommended by the ISCT are too basic and non-specific and there is a need of more than the minimal criteria to define MSCs [36] .
A deeper understanding and thorough study of the differentiation potential of MSC from different sources is warranted to enable the application of MSCs for molecular medicine [29] , to use MSCs in various forms of cellular therapies as well as genetic tools [30] and to enhance its potential use in cell and gene therapy for a number of human debilitating diseases and genetic disorders [31] .
Contributions
CV carried out the analysis & wrote the manuscript. BT carried out the experiments and performed the microarray. DY helped in the analysis. QL developed some of the methods used in the analysis as well as carried out initial analysis of hES-MSCs & BM-MSCs microarray data. VT conceptualized & designed the study, interpreted the results and supervised the study.
Conflict of Interest
The authors declare: “No competing financial interests exist”.
Acknowledgements
We would like to thank BII, A*STAR for funding this work. We would also like to thank Prof. Lim Sai Kiang, Institute of Medical Biology and Dr. Andre Choo, Bioprocessing Technology Institute for providing the hES-MSCs and Flb-MSCs.
Cite this paper
Vaz, C., Tee, B.T.B., Yong, D., Lee, Q.Y. and Tanavde, V. (2016) Mesenchymal Stromal Cells Derived from Human Embryonic Stem Cells, Fetal Limb and Bone Marrow Share a Common Phenotype but Are Transcriptionally and Biologically Different. Stem Cell Discovery, 6, 1-26. https://doi.org/10.4236/scd.2017.71001
References
- 1. Caplan, A.I. (1991) Mesenchymal Stem Cells. Journal of Orthopaedic Research, 9, 641-650.
https://doi.org/10.1002/jor.1100090504 - 2. Prockop, D.J. (1997) Marrow Stromal Cells as Stem Cells for Nonhematopoietic Tissues. Science, 276, 71-74. https://doi.org/10.1126/science.276.5309.71
- 3. Bianco, P., Riminucci, M., Gronthos, S. and Robey, P.G. (2001) Bone Marrow Stromal Stem Cells: Nature, Biology, and Potential Applications. Stem Cells, 19, 180-192.
https://doi.org/10.1634/stemcells.19-3-180 - 4. Minguell, J.J., Erices, A. and Conget, P. (2001) Mesenchymal Stem Cells. Experimental Biology and Medicine, 226, 507-520.
- 5. Horwitz, E.M., Le Blanc, K., Dominici, M., Mueller, I., Slaper-Cortenbach, I., Marini, F.C., Deans, R.J., Krause, D.S. and Keating, A. (2005) Clarification of the Nomenclature for MSC: The International Society for Cellular Therapy Position Statement. Cytotherapy, 7, 393-395.
https://doi.org/10.1080/14653240500319234 - 6. Caplan, A.I. (1994) The Mesengenic Process. Clinics in Plastic Surgery, 21, 429-435.
- 7. Pittenger, M.F., Mackay, A.M., Beck, S.C., Jaiswal, R.K., Douglas, R., Mosca, J.D., Moorman, M.A., Simonetti, D.W., Craig, S. and Marshak, D.R. (1999) Multilineage Potential of Adult Human Mesenchymal Stem Cells. Science, 284, 143-147.
https://doi.org/10.1126/science.284.5411.143 - 8. Krebsbach, P.H., Kuznetsov, S.A., Bianco, P. and Robey, P.G. (1999) Bone Marrow Stromal Cells: Characterization and Clinical Application. Critical Reviews in Oral Biology & Medicine, 10, 165-181. https://doi.org/10.1177/10454411990100020401
- 9. Zuk, P.A., Zhu, M., Mizuno, H., Huang, J., Futrell, J.W., Katz, A.J., Benhaim, P., Lorenz, H.P. and Hedrick, M.H. (2001) Multilineage Cells from Human Adipose Tissue: Implications for Cell-Based Therapies. Tissue Eng, 7, 211-228.
https://doi.org/10.1089/107632701300062859 - 10. De Bari, C., Dell’Accio, F., Tylzanowski, P. and Luyten, F.P. (2001) Multipotent Mesenchymal Stem Cells from Adult Human Synovial Membrane. Arthritis & Rheumatology, 44, 1928-1942.
https://doi.org/10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P - 11. De Bari, C., Dell’Accio, F. and Luyten, F.P. (2001) Human Periosteum-Derived Cells Maintain Phenotypic Stability and Chondrogenic Potential throughout Expansion Regardless of Donor Age. Arthritis & Rheumatology, 44, 85-95.
https://doi.org/10.1002/1529-0131(200101)44:1<85::AID-ANR12>3.0.CO;2-6 - 12. Maxim, M.A., Soritau, O., Baciut, M., Bran, S. and Baciut, G. (2015) The Role of Dental Stem Cells in Regeneration. Clujul Medical, 88, 479-482.
https://doi.org/10.15386/cjmed-475 - 13. O’Donoghue, K. and Chan, J. (2006) Human Fetal Mesenchymal Stem Cells. Current Stem Cell Research & Therapy, 1, 371-386.
https://doi.org/10.2174/157488806778226768 - 14. In’t Anker, P.S., Scherjon, S.A., Kleijburg-van der Keur, C., Noort, W.A., Claas, F.H., Wil-lemze, R., Fibbe, W.E. and Kanhai, H.H. (2003) Amniotic Fluid as a Novel Source of Mesenchymal Stem Cells for Therapeutic Transplantation. Blood, 102, 1548-1549. https://doi.org/10.1182/blood-2003-04-1291
- 15. Kim, D.W., Staples, M., Shinozuka, K., Pantcheva, P., Kang, S.D. and Borlongan, C.V. (2013) Wharton’s Jelly-Derived Mesenchymal Stem Cells: Phenotypic Characterization and Optimizing Their Therapeutic Potential for Clinical Applications. International Journal of Molecular Sciences, 14, 11692-11712.
https://doi.org/10.3390/ijms140611692 - 16. Lee, O.K., Kuo, T.K., Chen, W.M., Lee, K.D., Hsieh, S.L. and Chen, T.H. (2004) Isolation of Multipotent Mesenchymal Stem Cells from Umbilical Cord Blood. Blood, 103, 1669-1675. https://doi.org/10.1182/blood-2003-05-1670
- 17. Via, A.G., Frizziero, A. and Oliva, F. (2012) Biological Properties of Mesenchymal Stem Cells from Different Sources. Muscle, Ligaments and Tendons Journal, 2, 154-162.
- 18. Mareschi, K., Ferrero, I., Rustichelli, D., Aschero, S., Gammaitoni, L., Aglietta, M., Madon, E. and Fagioli, F. (2006) Expansion of Mesenchymal Stem Cells Isolated from Pediatric and Adult Donor Bone Marrow. Journal of Cellular Biochemistry, 97, 744-754.
https://doi.org/10.1002/jcb.20681 - 19. Sun, L.Y., Hsieh, D.K., Syu, W.S., Li, Y.S., Chiu, H.T. and Chiou, T.W. (2010) Cell Proliferation of Human Bone Marrow Mesenchymal Stem Cells on Biodegradable Microcarriers Enhances in Vitro Differentiation Potential. Cell Proliferation, 43, 445-456. https://doi.org/10.1111/j.1365-2184.2010.00694.x
- 20. Yoshimura, H., Muneta, T., Nimura, A., Yokoyama, A., Koga, H. and Sekiya, I. (2007) Comparison of Rat Mesenchymal Stem Cells Derived from Bone Marrow, Synovium, Periosteum, Adipose Tissue, and Muscle. Cell and Tissue Research, 327, 449-462.
https://doi.org/10.1007/s00441-006-0308-z - 21. Fraser, J.K., Wulur, I., Alfonso, Z. and Hedrick, M.H. (2006) Fat Tissue: An Underappreciated Source of Stem Cells for Biotechnology. Trends in Biotechnology, 24, 150-154.
- 22. Hass, R., Kasper, C., Bohm, S. and Jacobs, R. (2011) Different Populations and Sources of Human Mesenchymal Stem Cells (MSC): A Comparison of Adult and Neonatal Tissue-Derived MSC. Cell Communication and Signaling, 9, 12.
https://doi.org/10.1186/1478-811x-9-12 - 23. Cavallo, C., Cuomo, C., Fantini, S., Ricci, F., Tazzari, P. L., Lucarelli, E., Donati, D., Facchini, A., Lisignoli, G., Fornasari, P.M., Grigolo, B. and Moroni, L. (2011) Comparison of Alternative Mesenchymal Stem Cell Sources for Cell Banking and Musculoskeletal Advanced Therapies. Journal of Cellular Biochemistry, 112, 1418-1430.
https://doi.org/10.1002/jcb.23058 - 24. Jin, H.J., Bae, Y.K., Kim, M., Kwon, S.J., Jeon, H.B., Choi, S.J., Kim, S.W., Yang, Y.S., Oh, W. and Chang, J.W. (2013) Comparative Analysis of Human Mesenchymal Stem Cells from Bone Marrow, Adipose Tissue, and Umbilical Cord Blood as Sources of Cell Therapy. International Journal of Molecular Sciences, 14, 17986-18001.
https://doi.org/10.3390/ijms140917986 - 25. Liu, T.M., Martina, M., Hutmacher, D.W., Hui, J.H., Lee, E.H. and Lim, B. (2007) Identification of Common Pathways Mediating Differentiation of Bone Marrow and Adipose Tissue-Derived Human Mesenchymal Stem Cells into Three Mesenchymal Lineages. Stem Cells, 25, 750-760. https://doi.org/10.1634/stemcells.2006-0394
- 26. Wegmeyer, H., Broske, A.M., Leddin, M., Kuentzer, K., Nisslbeck, A.K., Hupfeld, J., Wiech-mann, K., Kuhlen, J., von Schwerin, C., Stein, C., Knothe, S., Funk, J., Huss, R. and Neubauer, M. (2013) Mesenchymal Stromal Cell Characteristics Vary Depending on Their Origin. Stem Cells and Development, 22, 2606-2618.
https://doi.org/10.1089/scd.2013.0016 - 27. Djouad, F., Bony, C., Haupl, T., Uze, G., Lahlou, N., Louis-Plence, P., Apparailly, F., Canovas, F., Reme, T., Sany, J., Jorgensen, C. and Noel, D. (2005) Transcriptional Profiles Discriminate Bone Marrow-Derived and Synovium-Derived Mesenchymal Stem Cells. Arthritis Research & Therapy, 7, R1304-R1315.
https://doi.org/10.1186/ar1827 - 28. Wang, S., Qu, X. and Zhao, R.C. (2012) Clinical Applications of Mesenchymal Stem Cells. Journal of Hematology & Oncology, 5, 19.
https://doi.org/10.1186/1756-8722-5-19 - 29. Caplan, A.I. and Bruder, S.P. (2001) Mesenchymal Stem Cells: Building Blocks for Molecular Medicine in the 21st Century. Trends in Molecular Medicine, 7, 259-264.
- 30. Reiser, J., Zhang, X.Y., Hemenway, C.S., Mondal, D., Pradhan, L. and La Russa, V.F. (2005) Potential of Mesenchymal Stem Cells in Gene Therapy Approaches for Inherited and Acquired Diseases. Expert Opinion on Biological Therapy, 5, 1571-1584.
https://doi.org/10.1517/14712598.5.12.1571 - 31. Patel, D.M., Shah, J. and Srivastava, A.S. (2013) Therapeutic Potential of Mesenchymal Stem Cells in Regenerative Medicine. Stem Cells International, 2013, Article ID: 496218.
https://doi.org/10.1155/2013/496218 - 32. Lai, R.C., Choo, A. and Lim, S.K. (2011) Derivation and Characterization of Human ESC-Derived Mesenchymal Stem Cells. Methods in Molecular Biology, 698, 141-150.
https://doi.org/10.1007/978-1-60761-999-4_11 - 33. Lai, R.C., Arslan, F., Tan, S.S., Tan, B., Choo, A., Lee, M.M., Chen, T.S., Teh, B.J., Eng, J.K., Sidik, H., Tanavde, V., Hwang, W.S., Lee, C.N., El Oakley, R.M., Pasterkamp, G., de Kleijn, D.P., Tan, K.H. and Lim, S.K. (2010) Derivation and Characterization of Human Fetal MSCs: An Alternative Cell Source for Large-Scale Production of Cardioprotective Microparticles. Journal of Molecular and Cellular Cardiology, 48, 1215-1224.
- 34. Ng, F., Boucher, S., Koh, S., Sastry, K.S., Chase, L., Lakshmipathy, U., Choong, C., Yang, Z., Vemuri, M.C., Rao, M.S. and Tanavde, V. (2008) PDGF, TGF-Beta, and FGF Signaling Is Important for Differentiation and Growth of Mesenchymal Stem Cells (MSCs): Transcriptional Profiling Can Identify Markers and Signaling Pathways Important in Differentiation of MSCs into Adipogenic, Chondrogenic, and Osteogenic Lineages. Blood, 112, 295-307.
https://doi.org/10.1182/blood-2007-07-103697 - 35. Brown, P.T., Squire, M.W. and Li, W.J. (2014) Characterization and Evaluation of Mesenchymal Stem Cells Derived from Human Embryonic Stem Cells and Bone Marrow. Cell Tissue Research, 358, 149-164.
https://doi.org/10.1007/s00441-014-1926-5 - 36. Tanavde, V., Vaz, C., Rao, M.S., Vemuri, M.C. and Pochampally, R.R. (2015) Research Using Mesenchymal Stem/Stromal Cells: Quality Metric towards Developing a Reference Material. Cytotherapy, 17, 1169-1177.
Supplementary Information
Supplementary File 1: Details of the differentially expressed genes (p-value, ratio, fold change) of hES derived MSCs.
The excel file comprises of 6 sheets namely:
Sheet 1: Adipocytes Day 7, Sheet 2: Adipocytes Day 21, Sheet 3: Chondrocytes Day 7, Sheet 4: Chondrocytes Day 21, Sheet 5: Osteocytes Day 7, Sheet 6: Osteocytes Day 21.
Supplementary File 2: Details of the differentially expressed genes (p-value, ratio, fold change) of Flb MSCs.
The excel file comprises of 6 sheets namely:
Sheet 1: Adipocytes Day 7, Sheet 2: Adipocytes Day 14, Sheet 3: Chondrocytes Day 7, Sheet 4: Chondrocytes Day 14, Sheet 5: Osteocytes Day 7, Sheet 6: Osteocytes Day 14.
Supplementary File 3: Details of the differentially expressed genes (p-value, ratio, fold change) of BM MSCs.
The excel file comprises of 6 sheets namely:
Sheet 1: Adipocytes Day 7, Sheet 2: Adipocytes Day 14, Sheet 3: Chondrocytes Day 7, Sheet 4: Chondrocytes Day 14, Sheet 5: Osteocytes Day 7, Sheet 6: Osteocytes Day 14.
Supplementary File 4: Comparison of significant canonical pathways among the three sources showing the canonical pathways commonly significant among all the three sources, among any two sources and specific to each source.
The excel file comprises of 6 sheets namely:
Sheet 1: Adipocytes Day 7, Sheet 2: Adipocytes Day 14/21, Sheet 3: Chondrocytes Day 7, Sheet 4: Chondrocytes Day 14/21, Sheet 5: Osteocytes Day 7, Sheet 6: Osteocytes Day 14/21.

Submit or recommend next manuscript to SCIRP and we will provide best service for you:
Accepting pre-submission inquiries through Email, Facebook, LinkedIn, Twitter, etc.
A wide selection of journals (inclusive of 9 subjects, more than 200 journals)
Providing 24-hour high-quality service
User-friendly online submission system
Fair and swift peer-review system
Efficient typesetting and proofreading procedure
Display of the result of downloads and visits, as well as the number of cited articles
Maximum dissemination of your research work
Submit your manuscript at: http://papersubmission.scirp.org/
Or contact scd@scirp.org